Journal of
eISSN: 2469 - 2786


Research Article Volume 10 Issue 3
St. Paul’s Inter College, India
Correspondence: Mohd Asjad Beg, St. Paul’s Inter College, Shahjahanpur, Uttar Pradesh -242001, India
Received: November 01, 2022 | Published: November 10, 2022
Citation: Baig MA. Hope toward potent drug target using tertiary topological instances of Mycobacterial transmembrane protein. J Bacteriol Mycol Open Access. 2022;10(3):69-75. DOI: 10.15406/jbmoa.2022.10.00330
Integral membrane proteins are important for various cellular processes. They are the major part involved in the transportation of different proteins across the membrane and thus might also be helpful in maintaining the charge potential. Rv0882 was predicted to be an integral membrane protein by various bioinformatics analyses. TBpred servers characterize the presence of this protein in the transmembrane region. SOSUI, TMHMM and HMMTOP servers found various transmembrane regions of this protein at various positions. TOPCONS web server provides the presence of an alpha-helical region in this protein. Secondary structure prediction and Three-dimensional (3D) modeling of this protein were done by using PSIPRED and QUARK respectively. Prediction of ligand binding site shows that various residues of this protein bind to different compounds whose results are confirmed by molecular docking of these compounds with Rv0882 protein. This protein also comprises a Fibronectin binding motif (RWFV) which also suggests that this protein might also act as a Fibronectin binding protein (FnBp). Previous studies proved that integral membrane proteins are important for transportation and FnBp are involved in host-pathogen interaction. Thus, studying this gene might be beneficial for stretching information about an untouched site of this bacterium which might be helpful in declining this infection.
Keywords: Mycobacterium tuberculosis, Rv0882, transmembrane protein, fibronectin binding-protein, pathogenesis
TB, Tuberculosis; M. tuberculosis, Mycobacterium tuberculosis H37Rv; M. africanum, Mycobacterium africanum; M. bovis, Mycobacterium bovis; M. microti, Mycobacterium microti; MTBC, Mycobacterium tuberculosis complex; FnBp, fibronectin binding proteins; PDB, protein data bank; TMHs, transmembrane helices; 3D, Three‐dimensional
Tuberculosis (TB) is a major threatening disease nowadays and is somewhere standing out of our control region. Majorly the causing agent behind this disaster is Mycobacterium tuberculosis H37Rv (M. tuberculosis), which infects humans and spreads by contacting an already infected patient.1 This pathogen comprises a complete complex known as the Mycobacterium tuberculosis complex (MTBC). This complex encompasses Mycobacterium tuberculosis infects the lungs, causing pulmonary tuberculosis, but this disease can also occur in other parts of the body, which is known as extrapulmonary tuberculosis.2–4 The main feature of this bacterial infection is the formation of granuloma. It comprises for the most part the enlistment at the stage in mycobacterial containment in macrophages.5 Although in the main case, the granuloma demonstrates the way to compel the infectivity; a few bacilli can really get into this arrangement for quite a while in a quiescent state.6 For a few purposes, which are still doubtful that the mycobacterial cell will pass by the quiescent state in 10% of the inactively tainted people, get away from the granuloma and spread all through the body, along these lines offering to ascend to clinical symptoms, and are at last dispersed all through environment.7–10 Various features of this bacterium add to its virulent character, and many of them had remained unstudied.11 One of the important features of the pathogenesis of any prokaryotic organism is its integral membrane proteins which allow the exchange and transportation of substances between the two sides of the membrane.12 These proteins are permanently embedded in the biological membrane and are in direct contact with annular lipids which are mentioned as the lipids with straight contact with proteins embedded in the membrane.13 These proteins include transporters, linkers, channels, receptors, enzymes, structural membrane-anchoring domains, proteins implicated in the gathering and transduction of energy and proteins accountable for the adhesion of the cell.14 These proteins could be divided into two super families: the outer membrane β-barrel proteins and those membrane proteins that have transmembrane α-helices.15,16 Most of these proteins engaged with aim of incorporation into the membrane by the Sec protein translocation pathway.17,18 The major partition of this family gene comprises transmembrane proteins which cover the intact biological membrane. Integral polytopic proteins and bitopic proteins are the types of integral membrane proteins.19 These proteins instead of crossing the transmembrane are attached to one side of the membrane. Rv0882 might be one of the integral membrane proteins of M. tuberculosis.20 This protein comprises transmembrane helices and motifs essential for binding to Fibronectin. This gene can be designated Fibronectin binding proteins (FnBp) due to the presence of the FnBp recognition motif RWFV. One of the studies proves that the RWFV motif is essential for the Fibronectin binding function of FAP-A which is the Fibronectin attachment protein of Mycobacterium avium.21 FnBp plays a major role in helping in the attachment of the pathogen to the host cell.22 Previous studies have found that several bacteria including M. tuberculosis occupy definite adhesion proteins that bind to the adhesive component found on the surface of the host cell. An example of FnBp in M. tuberculosis is antigen 85 complex (which comprises proteins named 85A, 85B, and 85C) is a major class of secretory proteins. FnBp can bind with the extracellular matrix which is the first step in the initiation of infection.23
Multiple sequences alignment is used for generating alignments between two or more sequences which are done by Clustal Omega and sequence retrieval by using the Mycobrowser database (https://mycobrowser.epfl.ch).24,25 The multiple sequence alignment of Rv0882 of M. tuberculosis had been done with orthologous Mycobacterium bovis (Mb0906), Mycobacterium leprae (ML2138c), Mycobacterium marinum (MMAR_4650) and Mycobacterium smegmatis (MSMEG_5686).
Subcellular localization studies
The prediction of the protein localization is done by using the TBpred server (http://crdd.osdd.net/raghava/tbpred/); this server is work on the support vector machine learning method for predicting the Mycobacterial protein localization. TBpred server predicts the accuracy module of this protein which is 86.62% respectively.26
Transmembrane helix prediction
The study of transmembrane helices (TMHs) is important in the context of membrane protein analysis. Many algorithm-based tools, such as Phobius, SOSUI, TMHMM, and HMMTOP, are already available to evaluate TMHs prediction. Phobius (http://phobius.sbc.su.se/) sever predicted the transmembrane topology and signal peptides for using the FASTA format amino acid sequence of a query protein.27 The SOSUI server (http://harrier.nagahama-i-bio.ac.jp/sosui) was used to distinguish between membrane proteins and soluble proteins based on the query protein's amino acid sequence.28 TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/) uses a hidden Markov model algorithm to predict the likely location and orientation of transmembrane helices. TMHMM server predicted that it contains a transmembrane helix and outputs the sequence labeled as inside or outside.29 The HMMTOP server (http://www.enzim.hu/hmmtop/index.php) is an automatic server that predicts TMH localization and protein topology based on a protein's amino acid sequence.30
Prediction of Alpha-helical transmembrane
The most important group of transmembrane proteins is alpha-helical transmembrane proteins, which play an important structural division of membrane proteins that are involved in a variety of cellular functions. Alpha-helical transmembrane protein prediction fabricates further consistent results are consensus prediction methods.31 To envisage the topology of Alpha-helical transmembrane proteins by using the TOPCONS web server had been used. TOPCONS web server (http://topcons.cbr.su.se/pred/) is a very user-friendly server and it’s predicted slightly faster. This server predicts the signal peptides from the transmembrane region and differentiates between globular and membrane proteins.32
Prediction of Beta-barrel transmembrane
Transmembrane beta-barrels (TMBBs) are very significant structural group of membrane proteins and might be involved in various cellular functions. There are several tools which are predicting the topology of beta-barrel transmembrane proteins.33 PRED-TMBB web server (http://bioinformatics.biol.uoa.gr/PRED-TMBB/) based on a hidden Markov model. PRED-TMBB web server is very user-friendly you can submit the FASTA format amino acids sequence. This web server predicted the topology of a query protein and assigned a score indicating the likelihood that the protein is an outer membrane beta-barrel protein, as well as subsequent possibilities for transmembrane strand calculation and a graphical representation of the implicit position of the transmembrane strands with respect to the lipid bilayer.34
Secondary structure prediction
The secondary structure of a protein is determined by the amino acid at the given position accompanied by its neighboring partners. Secondary structure prediction clearly shows the form of protein that covered the region in different forms whether it comprises alpha helices or containing beta barrels which is the key aspect of the prediction of protein configuration. The secondary structure prediction was studied with the PSIPRED (http://bioinf.cs.ucl.ac.uk) online tool.35,36
Three-dimensional (3D) protein model prediction and validation
If the query protein has a PDB homolog with a known structure, modelling the query amino acid sequence will be simple.37 The Rv0882 sequence was obtained from the Mycobrowser database, and three-dimensional (3D) modelling was predicted by the ab-initio modelling server QUARK online server, which constructs a protein's 3D model from an amino acid sequence. This online server used Monte Carlo simulation to create a model from a small fragment 1-20 residue long under the guidance of an atomic-level knowledge-based force field. This server does not have a homologous template in the PDB library.38 The 3D structure was modelled (sequence length 1-94) and refined by the 3D refine server (http://sysbio.rnet.missouri.edu/3Drefine/). In the 3D refine server follow the two-step first optimization of H bonds and second energy minimization by the MESHI molecular modelling package.39 Overall the model validation was done by the Structure Analysis and Verification Server (SAVES; http://servicesn.mbi.ucla.edu/SAVES/) metaserver40–42 and protein quality estimation by using ProQ server which has LGscore and MaxSub which define the model is fairly good, very good and extremely good.43 One more step to validate the molecular modelling analysis of the protein disordered region by using the DisEMBLTM, IUPred2A, PONDR and folding, unfolding analysis by Fold Index.44 DisEMBLTM (http://dis.embl.de/cgiDict.py) is used for the prediction of disordered/unstructured regions within a protein sequence.45 The next one is IUPred2A (https://iupred2a.elte.hu/) is a combined web interface that allows identifying disordered protein regions using ANCHOR2.46 Using the VL-XT method, we predict intrinsic order/disorder from amino acid sequences using PONDR (http://www.pondr.com/cgi-bin/pondr.cgi). The sequence-structure relationships suggest that disorder is an encoded property, and the predictions strongly suggest that proteins in nature have the far more intrinsic disorder than those in the Protein Data Bank.47 In the three-dimensional structure, there are amino acids interacted with each other, known as folded protein which is known as the native state. Here the prediction of Rv0882 is the folded or unfolded protein by using Fold Index. Fold Index (https://fold.weizmann.ac.il/fldbin/findex) is a web tool which provides a single score for the entire sequence, predicting whether it is folded or not by Kyte/Doolittle scale.48
Prediction of ligand binding pocket
Prediction of an active pocket of our modelled protein was done by RaptorX online server (http//: raptorx.uchicago.edu). Before active site-specific molecular docking (binding analysis) the determination of the true pocket is essential. The binding pocket is the site in the protein where the small molecules (ligands) can reversibly bind. Only a few amino acid residues are responsible for binding with a ligand. However other amino acid residues of the protein are provided with the correct orientation and confirmation. Relative modelled quality was evaluated by p-value statistics. For alpha protein, a p-value of less than 10-3 is a good indicator, and for beta protein p-value of less than 10-4 is a good indicator.49–51
Molecular docking
Insilico binding analysis of the Rv0882 and selected ligand by Protein Data Bank (PDB ID: HEA, DMU, PGV and PEK) perform the molecular docking and predicted the bound confirmations and binding affinities of their interactions. Auto dock tool-1.5.6 is used for docking purposes.52–60 To identify the binding sites of Rv0882 and ligand (PDB ID: HEA, DMU, PGV and PEK) which visualize the structure by the PyMOL and the prediction of the interacting residues in between protein and ligand are shown by using the LigPlot+.61–64
The sequence alignment was done by Clustal Omega, a sequence of Rv0882 protein of M. tuberculosis showing the orthologous homology with the Mycobacterium species when it was aligned with several different Mycobacterium species as shown in Figure 1.

Figure 1 Multiple sequence alignment using Clustal omega: Multiple sequence alignment Rv0882 of Mycobacterium tuberculosis H37Rv results out the homology and comparing the different mycobacterium species.
Subcellular localization studies
TBpred server was used for determining the localization of the protein comprised of 94 amino acid residues by a selected amino acid composition based on the SVM approach which predicted integral membrane protein with a score of 4.1473229.
Transmembrane helix prediction
Transmembrane helix predicted by using the Phobius, SOSUI, TMHMM and HMMTOP servers. Phobius server result is showing the topology of the domain (TOPO_DOM) and transmembrane helices (TRANSMEM). TOPO_DOM region of the 1st-11th and 63rd-67th is non-cytoplasmic else 36th-41st and 92nd-94th are cytoplasmic. TRANSMEM region is in 12th-35th, 42nd-62nd and 68th-91st. SOSUI server analysis of the protein hydrophobicity, if it exists this server makes a label that residue of this protein as a transmembrane region. As in result portion of the SOSUI server describe the transmembrane region as 11th-33rd, 40th-62nd and 68th-90th. TMHMM sever a result shows that in Rv0882 protein, there were 3 transmembrane helices (TMHs) 15th-17th, 42nd-60th and 70th-92nd. The predicted sequence is outside region 1st-14th and 61st-69th, the inside labeled sequence is 38th-41st and 93rd-94th as shown in Figure 2. HMMTOP server result had been shown that there were 3 transmembrane helices on the 14th-33rd, 42nd-61st and 70th-91st amino acid residues the output of the total entropy of the model is 16.9988 and the best path of the entropy is 17.0004. The predicted transmembrane by using the Phobius, SOSUI, TMHMM and HMMTOP servers is shown in Table 1.
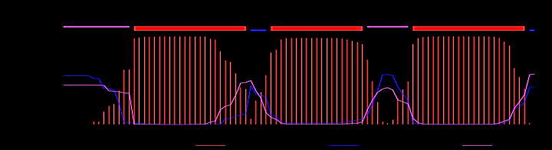
Figure 2 Transmembrane helix (TMHs) prediction: TMHMM server predicted the transmembrane helix (TMHs) region, in this Figure showing the TMHs region, are present in 15th-17th, 42nd-60th and 70th-92nd amino acid residues.
S.No. |
Phobius server |
TMHMM server |
SOSUI server |
HMMTOP server |
||
TOPO |
TRANSMEM |
outside |
TM helix |
TRANS. Region |
TRANS. Helices |
|
1 |
1-11 |
12-35 |
1-1 |
15-37 |
11-33 |
14-33 |
2 |
36-41 |
42-62 |
38-41 |
42-60 |
40-62 |
42-61 |
3 |
68-91 |
68-91 |
61-69 |
70-92 |
68-90 |
70-91 |
4 |
92-94 |
- |
93-94 |
- |
- |
- |
Table 1 Transmembrane helix prediction
Prediction of Alpha-Helical transmembrane
The TOPCONS web server is used to predict the most likely region of the membrane topology based on several individual algorithms. TOPCONS server predicted the given query of the FASTA format sequence using prediction algorithms (OCTOPUS, Philius, PolyPhobius, SCAMPI, and SPOCTOPUS). TOPCONS provided a result of the predicted transmembrane helix (TM) on TM1 (14th-34th), TM2 (42nd-62nd) and TM3 (71st-91st) as shown in Figure 3. Five of the other methods for prediction of alpha-helical transmembrane by using (OCTOPUS, Philius, PolyPhobius, SCAMPI and SPOCTOPUS) are shown in Table 2.
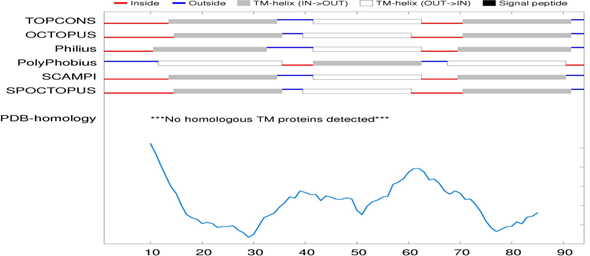
Figure 3 Predicted alpha-helical transmembrane: Alpha-helical transmembrane predicted by the TOPCONS web server results showing the predicted transmembrane helix (TM) on TM1 (14th-34th), TM2 (42nd-62nd) and TM3 (71st-91st).
S.No. |
TOPCONS |
OCTOPUS |
Philius |
PolyPhobius |
SCAMPI |
SPOCTOPUS |
1 |
14-34 |
15-35 |
11-32 |
12-35 |
14-34 |
15-35 |
2 |
42-62 |
40-60 |
42-62 |
42-62 |
42-62 |
40-60 |
3 |
71-91 |
71-91 |
70-91 |
68-90 |
70-90 |
71-91 |
Table 2 Prediction of Alpha Helical transmembrane
Prediction of Beta-Barrel transmembrane
PRED-TMBB web server predicted transmembrane beta-barrels (TMBBs) by different methods are the Viterbi method, N-best method and Posterior decoding method. The query sequence score is 2.788, which is lower than the threshold value of 2.965 this difference between the values indicates protein is in the outer membrane. PRED-TMBB web server result shows that protein region that was present in periplasmic space (in) 1st-61st, transmembrane strand (TM) 62nd-70th and 83rd-91st and extracellular space (out) 92nd-94th which is clearly shown in Figure 4.
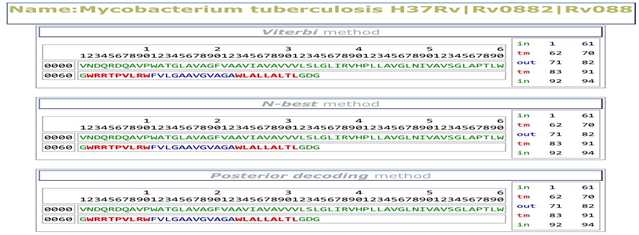
Figure 4 Prediction of Beta-Barrel transmembrane: PRED-TMBB server predicted the sequence is in periplasmic space (in) 1st-61st, transmembrane strand (TM) 62nd-70th and 83rd-91st and extracellular space (out) 92nd-94th.
Secondary structure prediction
PSIPRED predicted the secondary structure of the query protein Rv0882 of M. tuberculosis. predicted the Helix (H), extended strand in beta-sheet (E) and coiled region of the protein whereas the query protein has 74.46% of the α-helical region and coiled region 25.53% were the predicted event of 3 α-helices (15th-53rd, 59th-60th and 66th-92nd) rest residues region are coiled as shown in Figure 5.
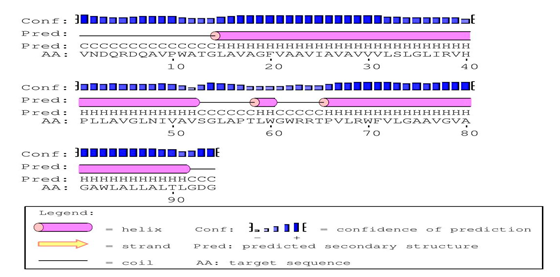
Figure 5 Secondary structure predictions: PSIPRED tool predicted the Secondary structure predicted has 3 α-helices extending from 15th-53rs, 59th-60th and 66th-92nd and the rest region is coiled.
Three-dimensional protein structure predictions and validation
3D structure of the Rv0882 of M. tuberculosis was modeled by the QUARK ab initio protein modeller which constructs a 3D model of correct fold 1/3 cases of short protein which have ≤200 amino acid residues. Modelled protein is refined by the 3DRefine and model validation is done by the SAVES metaserver, where are various parameters like Verify 3D, ERRAT score and Ramachandran Plot available in PROCHECK software. Modelled protein Rv0882 and Ramachandran plot analysis shown in this model 83.3% of residues are in an allowed region as shown in Figure 6. ERRAT score of the query protein is 98.8372 and the Verify 3D score is 71.28% which is acceptable for high-quality models. Protein quality prediction was done by the ProQ server results are LGscore is 5.214 and MaxSub is 0.312 which means this modelled protein has satisfactory confirmation. For predicting the disordered region by using the DisEMBLTM low-complexity predicted by CAST results shows all the amino acids are ordered and the IUPred2A results show the predicting structured proteins. At last, the PONDR web tool shows all the amino acids are in ordered form and the folding/unfolding analysis by using the Fold Index web tool which shows the Rv0882 protein sequence is folded which means this structure is in its native state as shown in Figure 7.
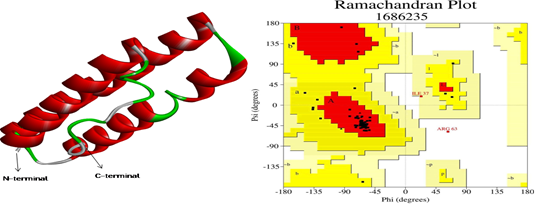
Figure 6 Predicted Model and RAMPAGE result: 3D structure of Rv0882 predicted by QUARK server and Ramachandran plot analysis shows 83.3% are the allowed region of the predicted model.

Figure 7 Prediction of ordered/disordered and folded/unfolded region of Rv0882 protein: In this figure (a) this subfigure clearly shows there are all the amino acids residues are in an ordered region which is done by using the PONDR web tool. (b) This subfigure shows, there are all residues are in the folded form which predicts this protein structure is in its native form.
Prediction of ligand binding pocket
Prediction of binding pocket was done by RaptorX; the query protein 5 pockets are predicted two best ligand HEME-A (HEA) and DECYL-BETA-D-MALTOPYRANOSIDE (DMU) with the multiplicity of 83 (HEA) and 53 (DMU). The parameter of pocket is true when the multiplicity of the pocket is above 40. The p-value of the modelled protein is 6.34e-03 which is less than 10-3 so they indicate the protein model is satisfactory for further evaluation. A description of the ligand of the binding pocket was shown in Table 3.
S.No. |
Multiplicity |
Ligand |
Binding residues |
|
1 |
1 |
83 |
HEA |
G14 L15 |
2 |
2 |
53 |
DMU |
N2 D3 Q4 |
3 |
3 |
42 |
PGV |
L32 G35 L36 R38 V39 L42 L43 L84 L87 |
4 |
4 |
32 |
PEK |
G35 V39 L91 G94 |
5 |
6 |
19 |
PEK |
V77 |
Table 3 Prediction of ligand binding pocket
Molecular docking
Molecular docking studies give precise orientations between receptors and compounds. The result of the docked conformations is selected on obtained energy parameters (binding affinities) and interacting residues as shown in Table 4. Docked conformations and interactions of the Rv0882 protein and selected ligand (PDB_ID HEA, DMU, PGV and PEK) are shown in Figure 8A and Figure 8B.
S. No. |
PDB ID |
Affinity (kcal/mol) |
Interacting partners |
1 |
HEA |
-6.4 |
Gln4, Asp6, Gln7, Ala8 |
2 |
DMU |
-5.6 |
Val1, Asp6, Gln7, Ala8, Ala22 |
3 |
PGV |
-4.7 |
Val1, Ala8 |
4 |
PEK |
-4.5 |
Asn2, Asp6, Gln7, Ala8 |
Table 4 Docking scoring and interacting partners generated from molecular docking showing the protein-ligand interaction
As we stand with an immediate urge to stop the pandemic of tuberculosis worldwide, various studies had been carried out to understand the physiology of the bacterium.22 This manuscript enlists one of that physiological understandings related to the integral membrane proteins. Integral membrane proteins are the major participator in the transportation process across the cellular membrane.35 This study found Rv0882 as a major integral membrane protein embedded in the plasma membrane of M. tuberculosis.11 Sequence alignment result shows that this protein is the homologue of other integral membrane proteins of Mycobacterium species as shown in Figure 1.26 Subcellular localization prediction shows its localization in the membrane portion as shown in Figure 2.27 Various transmembrane helix prediction servers predict transmembrane helices at various mentioned positions in this protein as shown in Figure 3.30 PRED-TMBB server shows that this protein is probably located in the outer membrane of the cell and periplasmic space at 1st-61st, transmembrane strands (TM) 62nd-70th and 83rd-91th and extracellular space (out) 92th-94th as shown in Figure 4.33 PSIPRED predicts the secondary structure of this protein by calculating the region covered by alpha helices, beta sheets and the coiled region as shown in Figure 5.35,36 The 3D structure of this protein had been sketched by QUARK ab-initio protein modeller and modelled protein was refined by 3D Refine as shown in Figure 6.37–39 Using DisEMBLTM, IUPred2A, PONDR and Fold Index showing Rv0882 amino acids sequence are ordered and folded, which means structures are in their native state.44–48 RaptorX was used for the predicted ligand binding site which shows that HEME-A (HEA) and DECYL-BETA-D-MALTOPYRANOSIDE (DMU) bind with the protein with the multiplicity of 83 (HEA) and 53 (DMU) as shown in Table 3.49 In the final step, molecular docking had been performed of the protein with mentioned ligand and confirmations have been selected because of energy parameters as shown in Figure 8A and 8B.52,53 Results of this study show that Rv0882 might be an integral membrane protein and help in transporting various materials across the plasma membrane and help in maintaining charge potential across the membrane. As we are in urgent urge to develop a treatment for this disease, this study might provide us with a better platform to understand the molecular pathology of this bacterium.
As the exposure to this disease increases day by day and we still need to reach the point of understanding the key mechanism of surviving this bacterium inside the host cell. Various aspects of different proteins of this bacterium had been studied in previous literature and are one of the important integral membrane proteins enlisted in this manuscript. Integral membrane proteins are attached to the transmembrane region of the cell of a bacterium and are important for transportation across the membrane. Rv0882 also consists of Fibronectin binding motif (FnBp) which also stretches our thinking towards the correlation between integral membrane proteins and Fibronectin binding proteins. This correlation might be helpful in pretending the role of these integral membrane proteins in pathogen attachment to the host cell. Understanding this mechanism thus takes us to a new track to abolish the mortality due to this malady.
It is my great pleasure to express my wholehearted gratitude to my St. Paul Inter College teachers. I really enjoyed the freedom she gave me to realize some of my ideas. I sincerely acknowledge Dr. Beg from Rutgers Cancer Institute of New Jersey, USA who supported me throughout, with their constructive annotations and treasured suggestions. Last but not the least, I would like to thank my family members as their endless love and solid support accompanied me all the time and without their Dua and Patience this article would not have been completed.
The author declares no conflicts of interest.

©2022 Baig. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.