Journal of
eISSN: 2469 - 2786


Research Article Volume 8 Issue 1
Centre for Interdisciplinary Research in Basic Science, Jamia Millia Islamia University, India
Correspondence: Fareeda Athar, Associate Professor, Centre for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia, New Delhi-110025, India
Received: April 29, 2020 | Published: June 9, 2020
Citation: Beg MA, Athar F. Pharmacokinetic and molecular docking studies of Achyranthes aspera phytocompounds to exploring potential anti-tuberculosis activity. J Bacteriol Mycol Open Access. 2020;8(1):18-27. DOI: 10.15406/jbmoa.2020.08.00268
Tuberculosis epidemic is at the highest risk therefore it becomes mandatory to define new defenses against it. Rapid securing of medication achievements of TB requests elective curative methodologies. Utilization of medicinal plants considered to fortify chemotherapeutic regimens. This study comprises of evaluation of 118 Mtb H37 Rv protein structures that were obtained from PDB databank and 19 phytocompounds of Achyranthes aspera L. that were selected through GC-MS analysis. The aim of the study is to define a portable anti-tuberculosis drug with lesser toxicity and higher efficacy. For fulfilling the demanded objectives, in silico methodology was applied. PyRx tool was used to prepare dock file and docking analysis was done by AutoDock Vina. Among 118 proteins, total 10 proteins were shortlisted based on the considering highest binding energy. Based on the top binding score, selected compounds were further analyzed by ADME properties to find the safe and effective drug-like compounds. The interaction studies were done by using Discovery Studio visualizer. Proteins with PDB IDs ITQ8, 1POH, 2FK8, 2JRC, 3B4Y, 3HZO, 3KXO, 3WQM, 4LJ1 and 5UGQ have high binding activity with compound number 1, 5, 10, 15, 16 and 17. The present study thus provide a perfect way to analyze the effect of phytocompounds on cumulative group of proteins that are predicted to be involved in essential processes of M. tuberculosis and thought to be better therapeutic target.
Keywords: Achyranthes aspera, ADMET, RO5, molecular docking, pharmacokinetics
XDR, extensively drug-resistant; EPTB, extra pulmonary tuberculosis; RRTB, rifampicin resistance tuberculosis; TST, tuberculin skin test; BCG, bacillus-calmette-guerin; NCBI, national centre for biotechnology information; PDB, protein data bank; MW, molecular weight; PDB, protein data bank; 3D, three dimensional; RO5, lipinski’s rule of five
Population of the world is at high risk of Tuberculosis (TB) which is caused by Mycobacterium tuberculosis H37 Rv, a highly professional pathogen that attacks on host very effectively.1 Primary infection of mycobacterium causes pulmonary tuberculosis, but it spreads to other parts of body and causes extra-pulmonary TB. Extra-pulmonary TB is commonly spreads in highly vascular areas such as kidney, eyes, lymph nodes, spine and bone.2 Although efforts has been made to reduce global challenge of drug-susceptible tuberculosis, the emergence of Multi-Drug-Resistant (MDR), Extensively-Drug-Resistant (XDR) and Total-Drug-Resistant (TDR) tuberculosis during the past decade threatens to destabilize these advances.3 MDR tuberculosis is caused by mycobacterium resistance at least to Isoniazid and Rifampicin, and XDR tuberculosis by mycobacterium resistance to Rifampicin and Isoniazid, any Fluoroquinolone, and one of the three injectable drugs, Capreomycin, Kanamycin, and Amikacin. TDR-TB is caused by Mycobacterium resistance to all first-line drugs and second-line drugs. Drug resistance severely threatens tuberculosis control, since it raises the possibility of a return to an era in which drugs are no longer effective.4 Drugs developed for treatment of MDR-TB and XDR-TB are bedaquiline and clofazimine but no drug is designed successfully to treat TDR-TB.5 Effective treatment against TB is very complex due to unique structure and composition of its cell wall because the complexity interferes with the entry of drugs into the organism. The most commonly known diagnostic test is simple skin test called the Mantoux test or Tuberculin skin test (TST).6 Bacillus-Calmette-Guerin (BCG) is the only vaccine used in TB prevention during early childhood.7 According to WHO Report of 2020, globally, In 2019, TB caused almost 1.5 million death among HIV people and there were an additional 251 000 deaths from TB among HIV-positive people. In 2019, 180 000 people developed TB that was Drug resistant. The united Nation (UN) meeting high-level meeting on TB provides a platform to step up the commitments and actions needed to end the global TB epidemic, by the SDG deadline of 2030.8 Due to the continuous development of resistant strains which pose the need for search and development of new drugs to cure this disease. For making new drugs medicinal plants are great hope which fulfil the need of treatment and it is very beneficial for human health.9-11 Achyranthes aspera plant is a well-known medicinal plant with roots, root bark, latex, leaves and flower parts which are used for various medicine to fight many human and animal diseases. A powerful spermicidal, antiparasitic, hypoglycemic, anticancer, anti-depressant, nephroprotective, anti-inflammatory, hepatoprotective, cardiovascular, bronchoprotective, anti-allergic, antioxidant, antibacterial, antifungal, and antituberculosis activity had been found to be present in various parts of A. aspera.12,13 The methanol extract of A. aspera were proved to be effective against the rifampicin induced hepatotoxicity in rats.14 A. aspera showed the presence of alkaloids, cardia glycosides, tannins, flavonoids, sterols and triterpenes.15 Earlier studies of the A. aspera plant reveals significant effect of different parts of plant in different pharmacological activities. According to the literature survey, more than eighty phytocompounds identified which involves in various activities.16 The present study suggests different A. aspera phytocompounds as inhibitors of A. aspera phytocompounds as an inhibitor which had examined by molecular docking method which predicted the novel hit and it could be a starting point for the development of more potent targeted compound having anti-tuberculosis activity.
Software and hardware
We used online databank PubChem (https://pubchem.ncbi.nlm.nih.gov/), Protein Data Bank (https://www.rcsb.org/), online tools such as Molinspiration (http://www.molinspiration.com//cgi-bin/properties), SwissADME (http://www.swissadme.ch/). Using software like ChemDraw, PyRxAutoDockTools-1.5.6 else using protein visualizer PyMOL and Discovery Studio.17–22 The schematic workflow of docking studies of A. aspera natural compounds against M. tuberculosis H37 Rv proteins as shown in Figure 1.
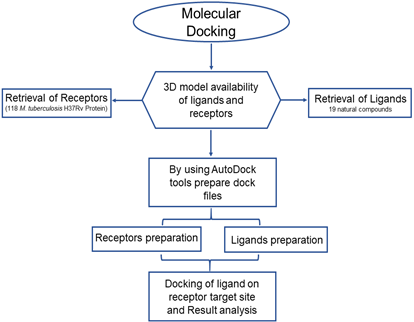
Figure 1 Schematic diagram of the workflow of docking studies on A. aspera natural compounds with M. tuberculosisproteins.
Preparation of the receptors ( tuberculosis proteins)
In our finding of natural compounds interacting effect of M. tuberculosis protein where the structure retrieve from PDB (protein data bank) online database, we have selected 118 protein of M. tuberculosis H37Rv based on categorization in functional manner. The first step is preparation of receptor which is necessary for carrying out binding studies, it can be specific (defined or site-specific molecular docking) or can be blind docking. Energy minimization is the necessary step for receptor preparation as it removes the poor contact in the protein structure and remove the conformational error in PDB structured protein. AutoDock Vina convert PDB to PDBQT which is followed by additional 3-4 steps such as delete water molecules, in hydrogen atoms add polar only and partial atomic charge were added to the macromolecule.18,19
Set dimension and generation of grid
In docking studies for the receptor (Protein), the first step is preparation of the grid by AutoDockTools-1.5.6. When the grid has been calculated then grid parameter file was saved in CONF file which is used for docking. In the auto grid procedure, the target protein is embedded into 3-dimension grid point.19
Preparation of the ligands ( aspera natural compounds)
In earlier studies, the GC-MS analysis identified the 19 phytocompounds of A. aspera which has highest peak, so we draw the 3D confirmation of these phytocompounds by using Molinspiration which is shown in Figure 2.16 Some of the natural compounds 3D structure was obtained from PubChem databases both are free databases and rest of it were drawn by using ChemDraw in SDF file which were converted to PDB using Open Babel GUI tool.17,18 There are 19 ligand (natural compounds) which were used for docking energy analysis and different pharmacokinetics analysis.16 Addition of partial charge, polar hydrogen, set rotatable bonds and checked missing atoms was done by AutoDockTools-1.5.6. Finally, the ligand became ready into PDBQT docking format.19
Pharmacokinetics analysis of ligand(s)
The Drug-Likeness properties of the A. aspera natural compounds were investigated by using the SwissADME server.23 SwissADME server is an online tool used for determining the drug like properties of the compound by uploading only SDF file then it first converted into Smiles which is followed by the command to run this file, so all the properties of the compound are on screen. The important molecular properties based on RO5 or Lipinski Rules of five, where 5 properties are seen are molecular weight (MW≤500 Dalton), number of hydrogen bond donor (H-bonds donor≤5), number of hydrogen bond acceptor (H-bonds acceptor≤10) and calculated Log P≤5.24,25 Prediction of the bioactivity scores was done by the Molinspiration and PASS which is an online server and support smiles file. It predicts the bioactivity score which is most important therapeutic targets like GPCR receptor, Kinase inhibitor, ion modulators, enzyme and nuclear receptor.26
Oral toxicity prediction
Present studies for clinical trial maximum drugs facing failure due to weak pharmacokinetics properties and cellular toxicity. Therefore, in silico pharmacokinetics profile of the natural’s compounds was evaluated by using ProTox-II online server. ProTox-II online server predicted the toxicity of the small compounds using smile format which is an important part of the drug design development process. The determination of the toxicity doses using computationally evaluation is too easy in comparison estimations to in animal models as it can reduce the time, amount of animal experiment.27
Molecular docking
Molecular docking studies were used for the finding of the protein inhibitor against the natural compounds of A. aspera plant. In this study using the ligand molecules (natural compounds of A. aspera plant) with receptor (118 M. tuberculosis H37 Rv protein) were conducted using AutoDocktools-1.5.6.19 Therefore, in this study I had shortlisted 10 proteins which involve in various process like- regulatory proteins; intermediary metabolism and metabolism; virulence, detoxification, adaptation; lipid metabolism; cell wall and cell processes, information pathway, PE-PPE and conserved hypothetical which is shown in Figure 3. By using blind or flexible docking the ligands set as the most feasible binding positions, on other hand the receptor was kept in defined state. The grid maps show the center of the active sites of the binding pocket for the ligand were calculated with Auto grid. After the docking there are 10 conformations were generated and we had selected the best-docked conformation based on the result of binding energy (Kcal/mol) for further analysis.
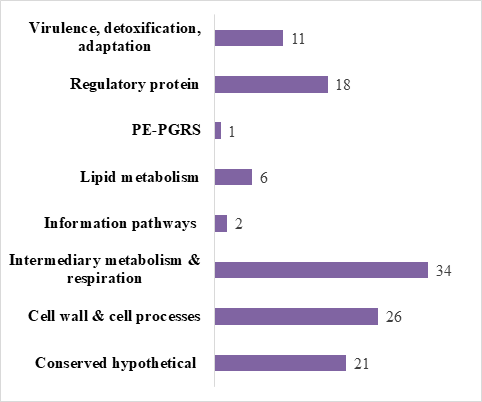
Figure 3In this figure showing the information of the receptor proteins for molecular docking analysis.
Visualization of the protein-ligand interaction
Binding interaction of the protein-ligand visualized among small ligand(s) and macromolecule was done by PyMOL software 1.3 version. PyMOL can produce high quality 3D image of small molecules and protein.20,21 The polar (hydrogen bond) and non-polar interactions between receptor and small ligand(s) were visualized by PyMOL software. For binding analysis of the 2D interaction, Discovery Studio 2019 was used .22
Pharmacokinetics analysis of ligand(s)
In 19 compounds from A. aspera plant, the drug-likeness properties are given in Table 1. These compounds were found to possess the best drug like properties by RO5.16 In A. aspera plant, natural compounds are of molecular weight in the range 102-466 (≤500). Accordingly, RO5 the low molecular weight of drugs is transported, diffuse and absorbed without any obstacle as compared to high molecular weight. Molecular weight (MW) as a significant characteristic of the in curative way of the drug action. In compound analysis RO5, next step is number of accepted hydrogen bond (O and N atoms) and number of donor’s hydrogen bond (NH and OH) within Lipinski’s limits range from 0-10 (H-bond acceptor) and 0-5 (H-bond donor) respectively.24 Lipophilicity (log P) and Topological Polar Surface Area (TPSA) values are crucial properties for the forecast of oral liability of drug molecules. The ranging of log P from (0-5) our most of the compounds range from 0.94-5.00 (≤5), which is the acceptable limits for drug to penetrate bio-membrane. The calculation of the surface areas which is occupied by oxygen, nitrogen and through attached hydrogen atom is called Topological Polar Surface Area (TSPA). Thus, the TPSA is meticulously associated to the hydrogen bonding potential of the compound.23 For a drug, a good bioavailability is more likely for compounds with ≤10 rotatable bonds and TPSA of ≤140Å. All the TPSA, miLogP, number of atoms, number of rotatable bonds and bioavailability is shown in Table 2.23 In analysis of A. aspera plant natural compounds, compound 9 and 12 have ≤10 number of rotatable bonds which is not good for bioavailability of the drug. All rest of the compounds (1-19) TPSA is 0.00 Å-138.44 Å range which is under 140Å and are in good bioavailability for drug like compound. Accordingly, there are all compounds except 9 and 12 have ≤10 number of rotatable bonds and flexible. All these compounds (1-19) TPSA range is in ≤140Å and that is good indicator for the oral route drugs.25 As a result, it was determined that the phytocompounds of Achyranthes aspera can be used as a natural better therapeutic target for antioxidant and antimicrobial resource.
|
S. No. |
Compound |
M.W. (g/mol) |
H-bonds acceptor |
H-bonds donor |
Log P |
RO5 |
|
1 |
17-PENTATRIACONTANOL(S) |
304.42 |
3 |
2 |
2.72 |
Y |
|
2 |
ACHRANTHINE(SE) |
129.16 |
3 |
1 |
1.17 |
Y |
|
3 |
ASARON(L) |
208.25 |
3 |
0 |
3.02 |
Y |
|
4 |
BETAINE(SE) |
102.15 |
1 |
0 |
-2.9 |
Y |
|
5 |
ECDYSONE(R) |
466.61 |
7 |
6 |
2.74 |
Y |
|
6 |
EUGENOL(L) |
164.2 |
2 |
1 |
1.66 |
Y |
|
7 |
HYDROQUINONE(L) |
110.11 |
2 |
2 |
0.92 |
Y |
|
8 |
NEROL(L) |
154.25 |
1 |
1 |
2.75 |
Y |
|
9 |
N-HEXACOS-14-ENOIC ACID(R) |
394.67 |
2 |
1 |
6.14 |
Y |
|
10 |
OLEANOLIC ACID(SE) |
470.73 |
3 |
2 |
3.95 |
Y |
|
11 |
P-BENZOQUINONE(L) |
108.09 |
2 |
0 |
0.94 |
Y |
|
12 |
PENTATRIAONTANE(S) |
492.95 |
0 |
0 |
9.16 |
Y |
|
13 |
SAPOGENIN(SE) |
414.62 |
3 |
1 |
4.43 |
Y |
|
14 |
SPATHULENOL(L) |
234.38 |
1 |
1 |
3.14 |
Y |
|
15 |
SPINASTEROL(S) |
414.71 |
1 |
1 |
5.05 |
Y |
|
16 |
ß-SITOSTEROL(S) |
400.68 |
1 |
1 |
4.91 |
Y |
|
17 |
STIGMASTA-5, 22-DIEN-3-OL(R) |
412 |
5 |
1 |
5 |
Y |
|
18 |
TETRACONTANOL-2(S) |
191.23 |
3 |
1 |
1.17 |
Y |
|
19 |
TRIACONTANOL(S) |
438.81 |
1 |
1 |
7.46 |
Y |
Table 1 Pharmacokinetics properties of natural compounds according to Lipinski rule analysis for A. aspera plant
|
S. No. |
Compound |
miLogP |
TPSA |
natoms |
nrotB |
nVio |
|
1 |
17-PENTATRIACONTANOL(S) |
2.07 |
57.53 |
22 |
1 |
0 |
|
2 |
ACHRANTHINE(SE) |
0.09 |
40.54 |
9 |
1 |
0 |
|
3 |
ASARON(L) |
2.49 |
27.7 |
15 |
4 |
0 |
|
4 |
BETAINE(SE) |
-3.81 |
17.07 |
7 |
2 |
0 |
|
5 |
ECDYSONE(R) |
1.11 |
138.44 |
33 |
5 |
1 |
|
6 |
EUGENOL(L) |
2.3 |
118.21 |
33 |
5 |
0 |
|
7 |
HYDROQUINONE(L) |
0.98 |
40.46 |
8 |
0 |
0 |
|
8 |
NEROL(L) |
3.2 |
20.23 |
11 |
4 |
0 |
|
9 |
N-HEXACOS-14-ENOIC ACID(R) |
9.53 |
37.3 |
28 |
23 |
1 |
|
10 |
OLEANOLIC ACID(SE) |
6.97 |
57.53 |
34 |
1 |
1 |
|
11 |
P-BENZOQUINONE(L) |
0.56 |
34.14 |
8 |
0 |
0 |
|
12 |
PENTATRIAONTANE(S) |
10.43 |
0 |
35 |
32 |
1 |
|
13 |
SAPOGENIN(SE) |
5.93 |
38.7 |
30 |
0 |
1 |
|
14 |
SPATHULENOL(L) |
4.15 |
20.23 |
17 |
0 |
0 |
|
15 |
SPINASTEROL(S) |
8.62 |
20.23 |
30 |
6 |
1 |
|
16 |
ß-SITOSTEROL(S) |
8.86 |
20.23 |
29 |
6 |
1 |
|
17 |
STIGMASTA-5, 22-DIEN-3-OL(R) |
7.87 |
20.23 |
30 |
5 |
1 |
|
18 |
TETRACONTANOL-2(S) |
0.1 |
49.66 |
14 |
4 |
1 |
|
19 |
TRIACONTANOL(S) |
9.98 |
20.23 |
31 |
28 |
1 |
Table 2 ADMET properties of natural compounds for A. aspera plant
*(R)Roots; (S)Stem; (L)Leaf; (SE)Seed; TPSA, Topological Polar Surface Area; natoms, number of atoms; nrotB, number of rotatable bonds; nVio, number of Violations
Bioactivity score prediction
The prediction of bioactivity score property of the 19 compounds with known inhibitors against GPCR receptor, Kinase inhibitor, ion modulators, protease inhibitor, nuclear receptor and enzyme activity were predicted by Molinspiration and PASS online server which is illustrated in Table 3.23,24 The acceptable range of the bioactivity score property 0.00 more active, -0.50 to 0.00 moderately active and <-0.50 less active. The result 19 compounds for A. aspera plant were biologically active and perform the physiological functions by interacting with GPCR receptor, Kinase inhibitor, ion channel modulators, protease inhibitor, nuclear receptor and enzyme activity. GPCR compounds are based on the signaling cascade which is used for the development of new functional drugs with better binding specificity and fewer unwanted effects. Through the bioactivity score result shows most of the compounds are in range 0.03 to -0.35 rest are above < -0.50 which compounds are 2, 3, 4, 7, 8 and 11 in the GPCR ligands. Next the ion channel modulators allow the movement of charge particle crosswise the cell membrane and are targeted by a collection of drugs. The result of the bioactivity score of ion channel modulators of A. aspera compounds was all the 0.02 to -0.43 rest are above < -0.50 which compounds are 2, 4, 7 and 11. Kinase inhibitor bioactivity score was 5, 6, 9, 10, 12, 17 and 19 are active further compounds score range is above < -0.50 which means those compounds are less active. Other inhibitor bioactivity score like kinase inhibitor, enzyme inhibitor and nuclear receptor ligand bioactivity score are 0.02 to -0.47 further score is above < -0.50 which means those compounds are less active.
|
S. No. |
Compound |
GPCR L |
Ion CM |
Kinase INH |
Nuclear RL |
Protease INH |
Enzyme INH |
|
1 |
17-PENTATRIACONTANOL(S) |
0.03 |
-0.28 |
-0.8 |
1.03 |
-0.12 |
0.68 |
|
2 |
ACHRANTHINE(SE) |
-1.5 |
-1.17 |
-2.62 |
-2.03 |
-1.85 |
-1.36 |
|
3 |
ASARON(L) |
-0.71 |
-0.43 |
-0.72 |
-0.47 |
-0.97 |
-0.39 |
|
4 |
BETAINE(SE) |
-2.96 |
-2.28 |
-3.68 |
-3.83 |
-3.4 |
-2.45 |
|
5 |
ECDYSONE(R) |
0.16 |
0.18 |
-0.32 |
0.87 |
0.3 |
0.66 |
|
6 |
EUGENOL(L) |
0.22 |
0.12 |
-0.35 |
1.06 |
0.33 |
0.67 |
|
7 |
HYDROQUINONE(L) |
-3.02 |
-2.48 |
-3.07 |
-2.84 |
-3.2 |
-2.66 |
|
8 |
NEROL(L) |
-0.6 |
0.07 |
-1.32 |
-0.2 |
-1.03 |
-0.28 |
|
9 |
N-HEXACOS-14-ENOIC ACID(R) |
0.18 |
0.05 |
-0.1 |
0.24 |
0.16 |
0.19 |
|
10 |
OLEANOLIC ACID(SE) |
0.26 |
0 |
-0.34 |
0.58 |
0.12 |
0.53 |
|
11 |
P-BENZOQUINONE(L) |
-3.54 |
-2.94 |
-3.17 |
-3.48 |
-3.51 |
-2.87 |
|
12 |
PENTATRIAONTANE(S) |
0.03 |
-0.01 |
-0.03 |
0.03 |
0.03 |
0.02 |
|
13 |
SAPOGENIN(SE) |
0.05 |
-0.14 |
-0.57 |
0.58 |
-0.06 |
0.61 |
|
14 |
SPATHULENOL(L) |
-0.35 |
-0.22 |
-0.61 |
0.19 |
-0.23 |
0.07 |
|
15 |
SPINASTEROL(S) |
0.14 |
0.04 |
-0.51 |
0.73 |
0.07 |
0.51 |
|
16 |
ß-SITOSTEROL(S) |
-0.02 |
0.18 |
-0.55 |
0.41 |
0.01 |
0.41 |
|
17 |
STIGMASTA-5, 22-DIEN-3-OL(R) |
0.12 |
-0.18 |
-0.48 |
0.74 |
-0.02 |
0.53 |
|
18 |
TETRACONTANOL-2(S) |
-0.46 |
-0.16 |
-1.03 |
-0.66 |
-0.23 |
-0.15 |
|
19 |
TRIACONTANOL(S) |
0.06 |
0.02 |
0.01 |
0.1 |
0.08 |
0.08 |
Table 3 Bioactivity Score of natural compounds for A. aspera plant.
*(R)Roots; (S)Stem; (L)Leaf; (SE)Seed. *GPCRL, G-Protein Coupled Receptor Ligand; Ion CM, Ion channel Modulator; Kinase INB, Kinase inhibitor; Nuclear RL, Nuclear receptor ligand; Protease INH, Protease inhibitor; Enzyme INH, Enzyme inhibitor.
Oral toxicity prediction
ProTox-II online server predicted the toxicity based on molecular similarity, fragment propensity and machine-learning program evaluated results is shown in Table 4.27
|
S. No. |
Compound |
LD50 (mg/kg) |
Toxic. Class (1-6) |
Avg. SM |
Pred. AC (%) |
MPSA |
|
1 |
17-PENTATRIACONTANOL(S) |
1860 |
4 |
97.62 |
72.9 |
57.53 |
|
2 |
ACHRANTHINE(SE) |
258 |
3 |
61.51 |
68.07 |
40.54 |
|
3 |
ASARON(L) |
418 |
4 |
100 |
100 |
27.69 |
|
4 |
BETAINE(SE) |
800 |
4 |
64.29 |
68.07 |
17.07 |
|
5 |
ECDYSONE(R) |
9000 |
6 |
100 |
100 |
138.45 |
|
6 |
EUGENOL(L) |
9000 |
6 |
100 |
100 |
118.22 |
|
7 |
HYDROQUINONE(L) |
225 |
3 |
100 |
100 |
40.46 |
|
8 |
NEROL(L) |
2100 |
5 |
100 |
100 |
20.23 |
|
9 |
N-HEXACOS-14-ENOIC ACID(R) |
48 |
2 |
100 |
100 |
37.3 |
|
10 |
OLEANOLIC ACID(SE) |
2000 |
4 |
100 |
100 |
57.33 |
|
11 |
P-BENZOQUINONE(L) |
25 |
2 |
100 |
100 |
34.14 |
|
12 |
PENTATRIAONTANE(S) |
154 |
3 |
100 |
100 |
0 |
|
13 |
SAPOGENIN(SE) |
8000 |
6 |
100 |
100 |
38.69 |
|
14 |
SPATHULENOL(L) |
3900 |
5 |
81.74 |
70.97 |
20.23 |
|
15 |
SPINASTEROL(S) |
890 |
4 |
91.26 |
72.9 |
20.23 |
|
16 |
ß-SITOSTEROL(S) |
667 |
4 |
91.86 |
72.9 |
20.23 |
|
17 |
STIGMASTA-5, 22-DIEN-3-OL(R) |
890 |
4 |
91.26 |
72.9 |
20.23 |
|
18 |
TETRACONTANOL-2(S) |
584 |
4 |
54.16 |
67.38 |
49.66 |
|
19 |
TRIACONTANOL(S) |
200 |
3 |
100 |
100 |
20.23 |
Table 4 Oral toxicity prediction of natural compounds for A. aspera plant
LD50, Lethal dose 50%; Toxic, Class- toxicity class; Avg. SM, Average similarity; Prediction accuracy and MPSA, Molecular Polar Surface Area.
Molecular docking
The molecular docking analysis of all 19 natural compounds of A. aspera plant accompanied by the flexible or blind docking method. This was used because of the finding inhibitor protein of M. tuberculosis H37Rv and each step and binding must be examined. AutoDocktools-1.5.6 (AutoDock Vina) tool was used to determine the molecular docking studies of the 118 number of M. tuberculosis H37Rv protein.19 Algorithm genetic process implemented in the AutoDock Vina was used. The molecular docking of ligands with 118 M. tuberculosis H37Rv proteins reveals that 10 proteins showed highest binding energy (Kcal/mol) which is shown in Table 5.19
S. No. |
Rv No |
PDB ID |
Binding affinity (Kcal/mol |
||||||
Lig 1 |
Lig 5 |
Lig 10 |
Lig 13 |
Lig 15 |
Lig 16 |
Lig 17 |
|||
1 |
1636c |
1TQ8 |
- |
-8.6 |
-10 |
-9.9 |
- |
-8.5 |
-8.9 |
2 |
819 |
1POH |
-9.4 |
- |
-11 |
-9.8 |
- |
- |
- |
3 |
0642c |
2FK8 |
- |
-9.2 |
-9.3 |
-9.8 |
- |
-9.7 |
- |
4 |
1014c |
2JRC |
- |
- |
-8.9 |
-10.2 |
-9.8 |
-9.6 |
-9.1 |
5 |
407 |
3B4Y |
-8.9 |
-9.2 |
-9.4 |
- |
- |
- |
-9.5 |
6 |
554 |
3HZO |
-9.7 |
- |
-9.4 |
- |
-9.1 |
-9.6 |
|
7 |
1364c |
3KXO |
-9 |
-9.3 |
-12 |
-12.3 |
-10.3 |
-9.9 |
-10 |
8 |
3378 |
3WQM |
- |
- |
-8.9 |
-10 |
- |
-9 |
-9.1 |
9 |
1566 |
4LJ1 |
- |
-9.2 |
-11 |
-10 |
- |
- |
- |
10 |
2224c |
5UGQ |
- |
- |
-11 |
-10.8 |
-9.1 |
-9 |
-9.3 |
Table 5 Ligand-receptor interaction of natural compounds which has highest binding affinity with M. tuberculosis H37Rv proteins.
*Rv No, M. tuberculosis H37Rv protein; Lig, ligand number; PDB ID, Protein Data Bank ID
Visualization of the protein-ligand interaction
After docking of the inhibiting ligands the protein-ligand interaction molecule, the ligands 1, 5, 10, 13, 15, 16 and 17 with proteins with PDB ID (1TQ8, 1POH, 2FK8, 2JRC, 3B4Y, 3HZO, 3KXO, 3WQM, 4LJ1 and 5UGQ) reveals that all the inhibitors binding active sites (means amino acid residues) occupy active pockets of the proteins as shown in Supplementary Figure 1 (A-J).22 In these studies of ligand-receptor interactions which is shown in Table 6 depicts, M. tuberculosis target protein and the type of interaction that is shown in between ligand-receptor from which resolve the important conundrum for which residues are substantial for protein stabilization and otherwise important residue that are important for protein conformation alteration.
S. No. |
PDB ID |
Ligand No. |
No. of Bonds |
No. of |
AA involved with interactive group |
H Bonds lengths (Å) |
1 |
1TQ8 |
5 |
5 |
3 |
HIS 44 (H); (O) |
2.71; 3.92 |
13 |
7 |
2 |
TRY 67 (O) |
2.95 |
||
16 |
6 |
1 |
TYR 40 (OH) |
2.25 |
||
2 |
1POH |
1 |
6 |
2 |
VAL 82 (OH) |
2.23 |
13 |
9 |
1 |
HIS 112 (OH) |
1.87 |
||
3 |
2FK8 |
5 |
7 |
5 |
ASP 26 (OH) |
1.88 |
10 |
7 |
1 |
ASP 26 (OH) |
2.27 |
||
16 |
7 |
1 |
ASP 26 (OH) |
2.51 |
||
4 |
2JRC |
10 |
5 |
1 |
ASN 23 (O) |
2.26 |
5 |
3B4Y |
1 |
8 |
3 |
THR 76 (O) |
2.97 |
5 |
6 |
3 |
GLU 13 (OH) |
2.10 |
||
10 |
5 |
1 |
THR 107 (O) |
2.80 |
||
6 |
3HZO |
10 |
7 |
1 |
ARG 21 (O) |
3.29 |
15 |
6 |
1 |
ARG 113 (O) |
113 |
||
16 |
5 |
1 |
CYS 186 (OH) |
2.18 |
||
7 |
3KXO |
All ligands have Pi-Alkyl bonds |
||||
8 |
3WQM |
All ligands have Pi-Alkyl bonds |
||||
9 |
4LJ1 |
5 |
6 |
2 |
SER 84 (O) |
3.18 |
10 |
8 |
2 |
SER 102 (OH) |
2.88 |
||
10 |
5UGQ |
13 |
6 |
1 |
ARG 388 (O) |
3.33 |
15 |
7 |
1 |
ARG 388 (O) |
2.82 |
||
Table 6 Ligand-receptor interaction with group involved in interaction of the receptor
*PDB ID, Protein Data Bank ID; H Bonds, Hydrogen bonds; AA, Amino acids, Å, Angstrom
Theoretically in molecular docking result in which 10 proteins with PDB ID (ITQ8, 1POH, 2FK8, 2JRC, 3B4Y, 3HZO, 3KXO, 3WQM, 4LJ1 and 5UGQ) have high binding activity with compound number 1, 5, 10, 15, 16 and 17. The result of type of bond present, distance between ligand and receptor and number of stability bonds have shown in Table 6. This in silico analysis is crucial for the protein selection for further in vitro analysis where the M. tuberculosis protein characterization and their evaluation as target for interaction with lead compound for future perspectives and final validation.28,29 Docking studies revealed that the PDB ID: 2FK8 interacted with the ligands 5, 10 and 16 but with ligand 5, it shows the highest Hydrogen group involved in interaction with receptor through amino acid residues ASP26, VAL27, TYR42, SER43, GLN108.
M. tuberculosis genome has been studied and decode over many decades ago but still there are many clues that remain untouched and may participate in bacterial resistance. This study identified phytocompounds against M. tuberculosis H37Rv proteins which are promising targets for developing therapeutics against the disease. Among all molecular docking studies result observations conclude that the selected ligands (natural compounds of A. aspera plant) were observed and selected 1, 5, 10, 15 16, 17 compounds as the best inhibitor for M. tuberculosis protein by considering six physicochemical parameters (LogP, TPSA, natoms, nrotB, nVio and MW) that were instituted to be important for the antituberculosis activity. Among all phytocompounds, 1, 5, 10, 15 16, 17 interacted with proteins was found to be with highest binding affinity and the interactive Hydrogen bonds with the highest dock score (−10.9) with compound 10. Targeted proteins were determined which will be helpful in further accentuate their mechanism of action in virulence and other essential pathways involved in survival and pathogenesis of the bacterium. In vitro work in this direction might provide some essential aspects of these proteins that make these proteins excellent target for approaching for therapeutic medication.
None.
The author declares that there is no conflicts of interest.

©2020 Beg, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.