eISSN: 2379-6367


Research Article Volume 8 Issue 4
Centre for Interdisciplinary Research in Basic Science, India
Correspondence: Dr Fareeda Athar, Associate Professor, Centre for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia, New Delhi-110025, India, Tel +91-11-26981717, Fax +91-11.26980164
Received: June 30, 2020 | Published: July 31, 2020
Citation: Beg MA, Thakur SC, Athar F. Molecular modeling and in silico characterization of mycobacterial Rv3101c and Rv3102c proteins: prerequisite molecular target in cell division. Pharm Pharmacol Int J. 2020;8(4):234-243. DOI: 10.15406/ppij.2020.08.00300
Mycobacterium tuberculosis H37Rv (M. tuberculosis) genetic evaluation found various conditional mutants that are being unsuccessful in surviving in non-permissive temperature or need enriched sources for growing in non-adaptive conditions. In M. tuberculosis basically, these mutants refer to as Filamentous Temperature Sensitive (FTS) gene family which have Rv3102c, Rv3610c, Rv2748c, Rv2151c, Rv2154c, Rv3101c, Rv2921c and Rv2150cproteins that are involved in major processes like cell division, cell growth and cell signalling.This manuscript provides a straightforward conduit towards finding the exact role of Rv3101c and Rv3102c genes in cellular cell divisionand survival of this bacterium by using computational approaches. The physiochemical properties shows both the proteins are stable and protein-protein interaction shows Rv2150c R2151c, smpB, Rv3104c and Rv2921c commonly interacted with each other but Rv3101c protein, a transmembrane protein which has four transmembrane helices which provide a clue for these proteins that act in various signalling cascades and protein exporting system. The protein phosphorylation play important role in several cellular processes and in Rv3101c phosphorylation occurs at Thr75, Ser89 and in Rv3102c phosphorylation occurs at residues Thr54, Thr166, Thr173, Ser174, Ser212 and Tyr225. The predicted structure was validated by SAVES metaserver and Ramachandran plot shows above 90% residues were present in the allowed region. In the prediction of B-cell and T-cell on Rv3101c and Rv3102c, protein antigen presentation and antibody binding properties were determined. These epitopes can be used to develop more effective vaccines and identify neutralizing antibodies. The mutation identification on the F126 and F1397 in Rv3101c whereas in Rv3102c on position F45 cause a drastic loss in inter-atomic interactions. As Rv3101c and Rv3102c proteins are essential class of proteins in M. tuberculosis, therefore, understanding of this class is crucial for analyzing cell division mechanism of this bacterium and thus help in clarifying its pathogenesis mechanism inside the host.
Keywords: Rv3101c, Rv3102c, cell division proteins, molecular modelling, structural stability
TB, tuberculosis; M. tuberculosis, Mycobacterium tuberculosis H37Rv; MDR-TB, multidrug-resistant TB; GTPases, Guanosine triphosphatases; G-proteins, GTP-binding proteins; SRP, signal recognition particle; FTS, filamentous temperature sensitive; PDB, protein data bank; NCBI, national center for biotechnology information; kD, kilodalton; THMs, transmembrane helices; 3D, three dimensional
Tuberculosis (TB), one of the ancient infectious diseases has now been transformed into an unreachable level. Although TB was described as pulmonary disease, manifestations of this disease have been increasing from lungs to all other parts of the body which is described as extrapulmonary tuberculosis (EPTB).1,2 Primary infection has been initiated by deposition of Mycobacterium tuberculosis H37Rv (M. tuberculosis), the known pathogen for this disease onto the surface of lung alveoli.3 The mortality rate due to this disease has been increasing through a much faster rate from last ten years.4 One of the major reasons behind this mortality and signature recognition for the pathogenesis of this disease is the formation of a granulomatous lesion in alveolar macrophages. Granuloma formation is generally based on inflammation of innate immune cellular components which is the result of the evolution of specified resistance of these bacilli towards host immunity.5-7 Granuloma provides mycobacterial cells with a cellular niche in which these cells can survive for a longer period without any interference of the host immune system. This phenomenon of granuloma formation leaves us with the conclusion that M. tuberculosis either leads to its manifestation to Tuberculosis (TB) or it can remain in the persistent stage which is growth-arrested dormant state.8 As this formation of granuloma is the result of many cellular and signal transduction processes it is very important to understand the mechanism to inhibit the initiation of the granulomatous lesion. This manuscript highlights one of the important families of genes which are responsible for cellular signal transduction mechanisms.9 Previous genetic evaluations identified several conditional lethal mutants which fail in surviving in non-permissive temperature or need enriched sources for growing in non-adaptive conditions. Communally, these genes termed as fts which can be designated as Filamentous Temperature Sensitive (FTS). FTS gene family is mainly responsible for cell growth and division of bacteria and involved in cell signalling. Previous work found essentiality of FTS family of genes in cell growth and division in various microorganisms including M. tuberculosis. In M. tuberculosis, FTS family comprises of Rv3102c, Rv3610c, Rv2748c, Rv2151c, Rv2154c, Rv3101c, Rv2921c and Rv2150c.10-16
In this protein family usually, most of the proteins are involved in the cell division, Rv2748c is a temperature sensitive protein in the native state rarely it involves in cell division. An assessment of the sequence homology of Rv2748c with Bacillus subtilis (B. subtilis) and M. tuberculosis, the B. subtilis sequence was much larger due to the presence of proline and glutamine (PQ) rich segments. More studies shows it might not to be essential for Escherichia coli (E. coli) but a significant one for reviewing the cell division and additional cellular mechanism of M. tuberculosis.17-19 For E. coli Rv2151c play a vital role in survival and were initially identified in experiments of temperature sensitive cell division, growth and septum formation.16 Rv2154cprotein leads a primary role in peptidoglycan synthesis of M. tuberculosis.17 In cell division process, Rv2150c protein act as a key player which involved in polymerization in GTP dependent manner like tubulin filaments and GTPases activity and starts the cell division process by gathering at the mid cell site to form inimitable structure called the Z-ring. In cell division Rv2150c and Rv2154c work in a supportive manner, this interface prophesied to be involved in the anchoring of Rv2150c to the membrane and involve this protein in septum formation.20,21
Rv2921found in operon which consist Rv3101c and Rv3102c and it is present in the inner membrane of the cell. The conditional lethal mutant of Rv3102c is an individual that divide in the nutrient enriched medium which remains the questionnaire sign for its direct involvement in the growth of a cell. Rv3102c is proven as an ABC transporter protein, which contains the nucleotide binding property and capacity of dimerization. This protein also participates in a variety of transport processes in prokaryotes as well as in eukaryotes. Rv3101c is also an integral membrane anchor protein work in combination with Rv3102c to form an ABC transporter cassette for conveying essentially salt across membrane.11,12,22
FTS gene family is majorly involved in cell division but also involved cell signalling pathways. This manuscript elaborates all FTS family genes that found in M. tuberculosis and their characteristics feature which helps for understanding the whole gene family. This manuscript also provides a straightforward conduit towards finding the exact role of these genes in cell survival of this bacterium. This Manuscript used in silico approaches to explore the Rv3101c and Rv3102c evident their biochemical and biophysical role in various cellular processes which empower virulence of this bacterium to see in Table 1.
|
S. No. |
Gene |
Length (amino acid) |
ATP/GTP binding site motif A |
ATP/GTP binding site motif B |
Gene ontology |
|
1 |
Rv3101c |
297 |
Absent |
159DRLFAVLD167 |
Cell division, Integral to membrane, Plasma membrane, Cell cycle |
|
2 |
Rv3102c |
229 |
36GPSGSGKS43 |
19DDINVKID27 163DEPTGNLD170 |
Atpase activity, Cell division, Atp binding |
Table 1 Enlistment of the proteins for Mycobacterium tuberculosis H37Rv which contain ATP binding site motif
Sequence retrieval and pertain physiochemical properties
To predict the physiochemical properties of Rv3101c and Rv3102c from Filamentous Temperature Sensitive (FTS) family protein of Mycobacterium tuberculosis H37Rv (Mtb) the amino acid sequence retrieves from the Mycobrowser database.23 The analysis of physiochemical features was predicted by ExPASy bioinformatics portal ProtParam tool which can compute the physical and chemical parameter for a given query amino acid sequence. This server evaluates the molecular weight, theoretical pI, amino acid composition, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index and grand average of hydropathicity (GRAVY).24,25
Protein-protein interaction
STRING database online server which was used to study the protein-protein interaction for many data repository sources for proteins into a single source which is easy to use. This server has executed the functionality to riddle out the interaction studies of proteins with a given source. STRING database interface consequence shown distinctly for each information and the cutoff value as low confidence: scores <0.4; medium: 0.4 to 0.7; high: >0.7.26,27
Alignment of transmembrane helices
Alpha-helical transmembrane proteins are the most important group of transmembrane proteins and play an important structural division of membrane proteins which implicated in a different array of cellular functions. Alpha-helical transmembrane protein prediction fabricates further consistent results are consensus prediction methods.28 To envisage the topology of Alpha-helical transmembrane proteins the TOPCONS web server was used. TOPCONS web server predicted slightly faster for the signal peptides from the transmembrane region and differentiates between globular and membrane proteins.29 Forinteractive protein data study was done by Protter it’s a web-based tool which supports to given visualizing hypothesis generation by using the amino acid sequence in the context of protein topology. Protter supports UniProt protein accession and numerous protein sequence, which can be readily extended via modular plugins. Protter visualizations sequence datasets show the integrated visual analysis of membrane proteins and peptide selection for targeted proteomics.30
Disorder enhanced phosphorylation
Phosphorylation of the proteins plays an important role in several cellular processes. The addition of phosphate group to a molecule is called Phosphorylation. When the phosphate group adds to an R group of an amino acid residue, it can change its conformation; hydrophobic side turns into a hydrophilic portion. Usually, the phosphorylation mediates the working and inhibition of many enzymes. It plays as an important component in transport, control and efficiency during glycolysis. Phosphorylation residues predicted by a DEPP (Disorder Enhanced Phosphorylation Predictor) server which predicted the phosphorylation site on the serine, threonine and tyrosine amino acid residues.31,32
Natural disordered region prediction
One more step to validate the protein topology analysis prediction of the disordered region done by PONDR online tool we predicted the intrinsic order/disorder from the amino acid sequence by VSL and VL3 method. The sequence-structure relationships indicate that disorder is an encoded property, and the predictions strongly suggest that proteins in nature are much richer in intrinsic disorder than are those in the Protein Data Bank.31,33
Molecular modelling
The main objective of the three-dimensional (3D) structure predictions from its amino acid sequence is to find out the best/accurate model achieved. There are various tools/online server which predicted the 3D structure of a protein-based homology modelling, fold recognition based and template-based modeling from its amino acid sequence. For, Rv3101c and Rv3102c protein modelling, we have used two online servers are Swiss-Model and Phyre2.34,35 SWISS-MODEL modelled the protein by the homology based which is fully automated. SWISS-MODEL build the protein model with the given targeted-template sequence alignment and Phyre2 suite which is based on Protein fold recognition.36-38 The 3D model of Rv3101c and Rv3102c protein generated from Swiss-Model and Phyre2 was compared and only the most suitable 3D model was selected for final validation.39 The final structures were validated the final 3D model by using RAMPAGE, Verify3D, ERRAT for stereo-chemical property.40-42
B-cell and T-cell epitopes
The Rv3101c and Rv3102c were designed to make target for antibodies with BepiPred 2.0 that identify likely binding sites (B-cell epitopes). We scan peptide sequences from individual protein with IEDB analysis resources for MHC-I and MHC-II presentation respectively to identify potential T-cell epitopes. We prophesied probable the human antibody binding sites (B-cell epitopes) on Rv3101c and Rv3102c protein with BepiPred 2.0.43 This study focused on neutralizing binding sites by only exploratory amino acids residues. For T-cell epitope Rv3101c and Rv3102c peptide prediction done by IEDB analysis server for MHC-I determination, we determined HLA-A*02:01 associated CD8 T-cell epitopes and 10mer sliding windows and MHC-II, we determined CD4 T-cell epitopes where specific HLA-DR alleles reported, so we used HLA-DRB1*09:01 and 15:01 (common alleles) we considered any peptide sequence above the 95th percentile as a positive epitope prediction.44-46
Mutation identification
In present, the prediction of the mutation analysis accurately done by various evolutionary tools and the mutational analysis also predicted by the physiochemical hypothesis that can predict and accurately measure the information created the exact and versatile Insilico way to deal with the mutation impact stability changes of the structure by sequence based prediction. This study plays a vital role to predict protein stability after a single point mutation. This is the way to deal with understanding the protein impacts of changes and connecting them with the ailment on the size of the entire proteome.47 In this study, we used EASE-MM server, which predicts amino acid substitutions in protein sequence and single nucleotide variants (SNVs). EASE-MM (Evolutionary, Amino acid, and Structural Encodings with Multiple Models) and its support vector machines applied to be a single domain monomeric protein model and predicted the (ΔΔGu) values. EASE-MM accomplishing a Pearson’s connection coefficient of up to 0.53-0.59 in 10 folds cross validation. By using the ProTherm, this server standardizes mutated stability of the protein which defines the difference in the Gibbs free energies of unfolding studies in between mutated and wild-type residues. If the ΔΔGu is less than 0 that means mutation decreases the protein stability. The ΔΔGu parameters of destabilizing protein: when ∆∆Gu is 1 then protein is stabilizing, 0.5 to -0.5 is neutral, -0.5 to -1 is likely destabilizing and more than -1 is destabilizing the protein.48,49 Structure stability changes upon point mutation were determined by the webserver, it may be utilized to study the effect of single point change on interaction level. The cut off worth set for this server is ΔΔG≥0 as stabilizing and ΔΔG<0 as destabilizing. Normal Mode Analysis (NMA) is a computational strategy that approximates the dynamic practicality of a structure around an adjustment through consonant development. This has been used to make possible improvements and appropriately give beneficial bits of information into protein developments, and their accessible conformational assortments.50
Sequence retrieval and pertain physiochemical properties
For retrieval of the sequence, we have used Mycobrowser database which provides the FASTA format sequence Rv3101c and Rv3102c protein of M. tuberculosis.23 Analysis of physiochemical properties was done by ExPASy server, ProtParam tool to predict the molecular mass (kD), Isoelectric point (IP), instability index, aliphatic index and GRAVY which is shown in Figure 1. In Rv3101c and Rv3102c protein, the physiochemical properties show both proteins are stable by Instability index.24,25
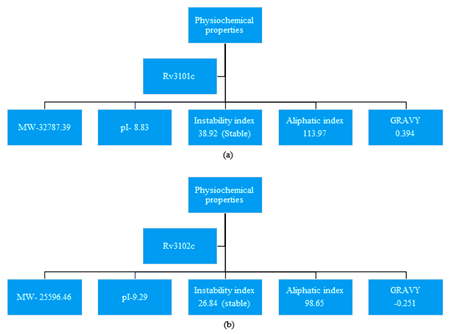
Figure 1 ProtParam tool to explore the physiochemical properties (a) Rv3101c and (b) Rv3102c from M. tuberculosis H37Rv.
Protein-protein interaction
The study of protein-protein interaction was done by the STRING database server which result from demonstration that Rv3101c and Rv3102c commonly interacted with the Rv2150c R2151c, smpB, Rv3104c and Rv2921c allboth protein interaction is shown in Figure 2. STRING database server produces the score built on an interacting protein which illustrate the cutoff value among the minimum interaction score (between 0.4 -0.6) and above score showed high interaction. Based on an interacting score Rv2150c (Cell division protein FtsZ; it is essential for the cell division that forms a contractile ring which is called Z ring at the future cell division site) protein shows the high interacting score i.e. 0.934 with Rv3101c and 0.981 with Rv2150c.26,27
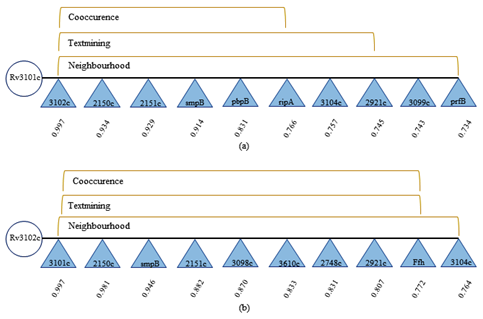
Figure 2 The protein-protein interaction was done by STRING server (a) R3101c and Rv3102c proteins where score shown separately for each data confirms cut off value within [0-1], low confidence: scores <0.4; medium: 0.4 to 0.7; high: >0.7.
Alignment of transmembrane helices
Alpha-helical transmembrane predicted by using the TOPCONS web server which concludes the most probable region of the membrane topology based on the several individual algorithms. TOPCONS server predicted given query of FASTA format sequence with (OCTOPUS, Philius, PolyPhobius, SCAMPI and SPOCTOPUS) prediction algorithms. TOPCONS provided a result of the predicted transmembrane helix (TM) on TM1 (22nd-42th), TM2 (172nd-192nd), TM3 (222nd-242th and 269th-289th) as shown in Figure 3. Five of the other methods for prediction of alpha-helical transmembrane was used (OCTOPUS, Philius, PolyPhobius, SCAMPI and SPOCTOPUS).28,29 The envisioning premise of transmembrane helices (TMH) by using amino acid for protein topology shows Rv3101c is a membrane protein which consists four TMHs on position 21st-45th, 171st-197th, 218th-246th and 266th-281th and Rv3102c has not found any TMH. These findings identify that Rv3101c and Rv3102c amino acid sequence has N-glyco motifs which are shown in Figure 3.30

Figure 3 Transmembrane helices prediction in Mycobacterial proteins (a) In Rv3101c protein has predicted four transmembrane helices (b) In Rv3102c were not found any transmembrane helices.
Disorder enhanced phosphorylation
Disordered phosphorylation, prediction was done by DEPP server. In Rv3101c the predictedphosphorylated residues are Thr75, Ser89 and in Rv3102c residues are Thr54, Thr166, Thr173, Ser174, Ser212 and Tyr225. The predicted phosphorylated Serine in Rv3101c – 89 out of 13 residues (7.6923%) and phosphorylated Threonine on position Thr75 out of 17 residues (5.8824%). In Rv3102c predictedphosphorylated sites, disorder phosphorylated Serineis 174 and 212 out of 12 (16.6667%), phosphorylated Threonine is 54, 166 and 173 out of 14 (21.4286%) and phosphorylated Tyrosine is 225 out of 3 (33.3333%) which is shown in Figure 4.31

Figure 4 Phosphorylation site prediction of Mycobacterial proteins (a) In Rv3101c has predicted phosphorylation site as 75 and 89 position residues (b) In Rv3102c has predicted phosphorylation sites on Thr54, Thr166, Thr173, Ser174, Ser212 and Tyr225 residues.
Natural disordered region prediction
In Rv3101c and Rv3102c, the natural disordered region prediction was done by PONDR by using VSL2 and VL3. In the comparison of this plots VSL2 and VL3 if the score above 0.5 then the region was predicted disordered and scores below 0.5 then the region was predicted ordered. In Rv3101c by using PONDR server the VSL2 plot has scores above 0.5 which predicted disordered region on 66-77, 82-89, 116-120, 132-142 and 295-297 residues and in Rv3102c VSL2 plot predicted disordered region on 1-15, 40-41, 56-60, 132-143, 165-175 and 218-229 and VL3 shows 220-229 residue region is disordered. Comparison of disorder residues prediction between PONDR-VSL2 and PONDR VL3 is shown in Figure 5.31,33

Figure 5 In natural disordered region prediction, comparison of graph plot between VSL2 and VL3plots, where scores greater than 0.5 are predictions of disordered residues. (a) In Rv3101c graph plot represent regions (from N- to C- termini) and (b) in Rv3102c graph plots the peaks above 0.5 represents disordered region.
Molecular modelling
The molecular modelling for Rv3101c and Rv3102c FTS family proteins of M. tuberculosis was done by using homology modeling by the SWISS-MODEL and fold recognition modelling was done by Phyre2 server.34,35,39 The primary criteria of the selected template are maximum identity, large alignment and fewer gaps. The steps involved to choose the best built 3D model prediction and the final model predicted by using (SWISS-MODEL and Phyre2 server) was comparatively analyzed based on protein validation which is done by SAVES metaserver (RAMPAGE, Verify3D and ERRAT servers) which is shown in Figure 6.40 In this comparative analysis for Rv3101c the best build model was made from Swiss-Model, that the given model has template PDB ID: 4N8N (Cell division protein) which has 100% sequence identity but the coverage sequence 45-157 residues, the model validation by Verify scored 100.00% (Pass), ERRAT score 95.6989 and the RAMPAGE server validates the protein structure based on φ, ψ angle of individual residues percentage of the most favoured region is 95.7%, So the protein quality prediction is satisfactory. For Rv3102c the best build model was made from Phyre2, that the given model has template PDB ID: 3DHW which has 100% confidence and 40% sequence identity and the model validation by Verify scored 94.76% (Pass), ERRAT score 79.638 and the φ, ψ angle of individual residues percentage of the most favoured region is 93.5%, a good quality model has expected over 90% of the residues in the most favoured region which is shown in Figure 6.41,42
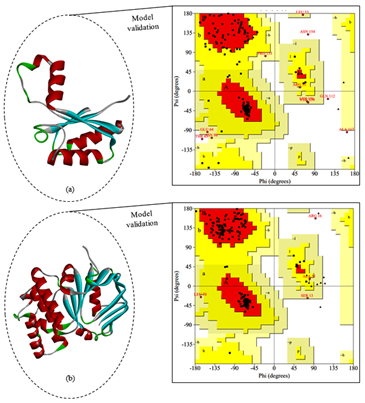
Figure 6 Molecular modelling was done by Swiss Model and Phyre2 (a) In cartoon model of Rv3101c predicted model and the plot shows 95.7% residues are in favoured region (b) In Rv3102c model predicted by Phyre2 and the plot shows 93.5% residues are in favoured region.
B-cell and T-cell epitopes
In Rv3101c and R3102c the prediction of B-cell epitope was done by BepiPred 2.0 online tool for target antibody binding sites. In Rv3101c the predicted peptide taking the overlapping window of 10 amino acid with the set threshold at 0.50 with 75% specificity. The predicted B-cell epitope were as 66VSANDSSCDTT76, 95RFLNRQQAYD104, 111PQFKDVAGKD120 and 149VLDVLNQKEL158. In Rv3102c the predicted B-cell epitope were as 67VNKLRGRHVP76, 109GKRTDAINRV118, 127GLSGKANRLP136 and 167GNLDPETSRD176.43 The T-cell epitope prediction was done by IEDB analysis (MHC I and MHC II) in Rv3101c the specific allele HLA-A*02:01 alleles associated CD8 T-cell predicted epitope as 163FAVLDGLSNA172, 23AMILTTAISV32 and 220ILTTAISVGL229 and in Rv3102c the MHC I T-cell predicted epitope as 33GLFGGGMLVV42, 156KELIDRLFAV165 and 32VGLFGGGMLV41. In MHC II multiple DR-β1 (DRB) alleles were used like HLA-DRB1*09:01 and 15:01 and we considered above 95 percent as 70DSSCDTTACKA80, 67SANDSSCDTTA77, 145GQPGVLDVLNQ155 and 69NDSSCDTTACK79 in Rv3102c the MHC II predicted peptide as 51AAETPTSGDVR61, 162ADEPTGNLDPE172, 15ALDDINVKIDK25 and 21DINVKIDKGEF31. All the predicted B-cell and T-cell epitope as shown in Figure 7.44-46

Figure 7 B-cell and T-cell epitope predicted residues for target antibody binding sites. (a) predicted peptide in Rv3101c and (b) predicted peptide in Rv3102c it comprises the protein secondary structure formation.
Mutation identification
The prediction of the mutation was done by the EASE-MM server. The protein stability change in residue by the sequence based method and DynaMut server predicted the mutated amino acid changes in protein structure regarding protein stability.47 EASE-MM server comprises of five specialized models of support vector machines and then selected final prediction was based on selected secondary structure model and its surface area. After EASE-MM calculations of mutations, selected residues in Rv3101c were Phenylalanine (F126), Phenylalanine (F1397) and in Rv3102c Phenylalanine (F45).48,49 Impact of the point mutation was also studied by using DynaMut server which had two diverse ways, (1) study of protein dynamics and (2) effect of point mutation on protein structural stability. In this server, mutation study includes three different servers mCSM, SDM and DUET and EASE-MM predicted selected highest destability for mutation for Glycine which has the highest negative score which is shown in Table 2. DynaMut server predicted the effect of interatomic interaction on the single amino acid mutation and predicted the drastic loss in Interatomic interactions. All hydrogen bonds, halogen bonds and hydrophobic interaction were disrupted as shown in Figure 8.50
|
S. No |
Online Server |
Rv3101c |
Rv3102c |
|
|
F126G |
F139G |
F45G |
||
|
1 |
∆∆G DynaMut |
-3.603 kcal/mol |
-3.206 kcal/mol |
-1.042 kcal/mol |
|
2 |
ΔΔG mCSM |
-3.280 kcal/mol |
-3.631 kcal/mol |
-1.703 kcal/mol |
|
3 |
ΔΔG SDM |
-2.330 kcal/mol |
-2.330 kcal/mol |
-2.330 kcal/mol |
|
4 |
ΔΔG DUET |
-3.655 kcal/mol |
-3.868 kcal/mol |
-1.954 kcal/mol |
|
5 |
∆∆G ENCoM |
-1.574 kcal/mol |
-1.097 kcal/mol |
-1. 040 kcal/mol |
|
6 |
ΔΔSVib ENCoM |
1.968 kcal.mol-1.K-1 |
1.371 kcal.mol-1.K-1 |
1.300 kcal.mol-1.K-1 |
Table 2 In Rv3101c and R3102c predicted mutation-induced stability changes by Structure-based analysis
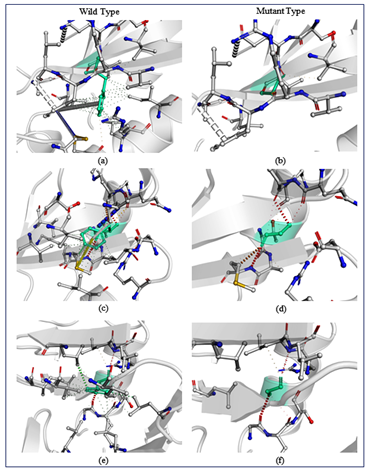
Figure 8 In mutation analysis on selected residues for visualizing the drastic changes in interatomic interaction between wildtype (WT) and mutant (MT) which shows mutation showing the bond disruptions (a-d) Rv3101c residues F126 and F139 mutated with Glycine predicted drastic changes in protein stability.(e-f) Rv3102c residues F45 mutated with Glycine predicted drastic changes in protein stability.
Tuberculosis has now become a major threat and it is almost become unattainable to cure this disease after applying so many methodologies. Decades of research have applied their methodologies to accomplish a better cure for this disease, but it seems unreachable. This unsuccessful treatment of this disease is due to its unusual nature of this bacterium to the magnifying mechanism of mutation in genome. We all are facing an unbalanced equation of solving tuberculosis. Although many kinds of research had been failed at some point, we are getting some successes also. We here present combined information regarding a very important family of genes present in M. tuberculosis. These studies proved that Rv3101c and Rv3102c proteins ATP binding and ABC transporter proteins which involve in cell growth and division of this bacterium but also participate in signal transduction and ribosome assembly. This manuscript thus provides information regarding Rv3101c and Rv3102c proteins in a concise way which provides a better way to understand this family and help in generating a universal treatment for this disease. Therefore, an outlook of these proteins is an essential step in understanding which might help in attaining an additional platform towards the eradication of this disease.
Md Amjad Beg: Conceptualization idea, Methodology designing, Software handling, writing manuscript and evaluate results Dr Fareeda Athar: Reviewing and corresponding author.
The authors acknowledge support from the Centre for Interdisciplinary Research in Basic Science, Jamia Millia Islamia University. Md Amjad Beg also acknowledges UGC-MANF (MANF-2017-18-UTT-88071) for the financial support and Jamia Millia Islamia University.
The authors declare that they have no potential conflict of interests.

©2020 Beg, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.