Journal of
eISSN: 2469 - 2786


Research Article Volume 8 Issue 3
Centre for Interdisciplinary Research in Basic Science, Jamia Millia Islamia University, India
Correspondence: Fareeda Athar, Associate Professor, Centre for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia University, New Delhi-110025, India
Received: September 03, 2020 | Published: September 18, 2020
Citation: Amjad Beg MD, Ansari S, Athar F. Molecular docking studies of Calotropis gigantea phytoconstituents against Staphylococcus aureus tyrosyl-tRNA synthetase protein. J Bacteriol Mycol Open Access. 2020;8(3):78-91. DOI: 10.15406/jbmoa.2020.08.00278
The current study reports Calotropis gigantea (C. gigantea) commonly known as milk weed contains phytoconstituents that have various medicinal properties. In Present study, Molecular docking analysis was also carried out to validate the anti-bacterial potential of these phytoconstituents. The reported previous studies of C. gigantea essential oil extract showed the GC-MS analysis and its screening found occurrence of nineteen compounds with different biological activity. The docking studies of C. gigantea phytoconstituents against S. aureus tyrosyl-tRNA synthetase (PDB ID: 1JIJ) found that it plays an essential role in protein synthesis by producing charged tRNAs. In docking analysis out of nineteen, there are seven compounds which shows highest docking score range -6.8 to -6.3 which are namely 2,4-dimethylacetophenone; 2-Methoxy-4-vinylguaiacol; 3-Phenyl-2-propenoic_acid; 5-Methyl-2,4-diisopropylphenol; Eugenol; α-Terpineol, L-Tyrosine_Methyl_ester. The docking analysis showed that all the nineteen compounds showed in range of -4.5 to -6.8 Kcal/mol docking energy. The L-Tyrosine_Methyl_ester docking energy -6.7, showing the maximum hydrogen bond interaction with TYR36, ASP40, ASP80 and GLN19. Thus, these studies might be beneficial for stretching information about an untouched site of this topic that will be helpful in declining this infection.
Keywords: Calotropis gigantea, Staphylococcus aureus, tyrosyl-tRNA synthetase, molecular docking
C. gigantea, Calotropis gigantea; S. aureus, Staphylococcus aureus; TyrRS, tyrosyl-tRNA synthetase; RO5, Lipinski’s rule of five; 3D, three‐dimensional; RB, rotatable bond; HBA, hydrogen bond acceptor; HBD, hydrogen bond acceptor; log P, lipophilicity; TSPA, topological polar surface area
Plants, which have at least one of its parts having substances that can be utilized for treatment of diseases, are called therapeutic plants.1 Drugs got from plants are generally acclaimed because of their wellbeing, simple accessibility and ease. Home grown meds may incorporate entire pieces of plant or generally set up from leaves, roots, bark, seed and blossoms of plants. Medicinal plants are the "spine" of customary medication, which implies more than 3.3 billion individuals in the less evolved nations.2,3 C. gigantea Linn is an enduring bush normally known as milkweed or wasteland weed. It has a place with family Asclepiadaceous and was assessed for its anticonvulsant movement. It has groups of waxy blossoms that are either white or lavender in shading.4 There is a constant need of the advancement of new compelling antimicrobial medications due to the development of new irresistible ailments and medication opposition.5,6 In Calotropis gigantea Linn. bloom constituent, for example, 2,3-Dihydro-benzofuran, 2,4-dimethylacetophenone, 2-Methoxy-4-vinylguaiacol, 3,7-Dimethyl-1,6-octadien-3-ol, 3,7-dimethyl-2,6-octadien-1-ol, 3-Phenyl-2-propenoic corrosive, 3-Thiophenemethanol, 5-Methyl-2,4-diisopropylphenol, 6-hepten-1-ol, 2-Methyl, Benzaldehyde, Benzyl liquor, Cinnamyl liquor, Eugenol, Furfural, α-Terpineol, L-Tyrosine Methyl ester, Phenethyl liquor, Pinocampheol, Trans-3-Hexen-1-ol.7 The plant is accounted for pain relieving action, antimicrobial movement, cell regeneration action, hostile to pyretic action, insecticidal action, cytotoxicity action, hepatoprotective action, pregnancy interceptive properties, laxative properties, pro coagulant action and wound mending action.8 ESKAPE is an abbreviation including the names of six bacterial microorganisms generally connected with antimicrobial opposition (Detection of ESKAPE Bacterial Pathogens at the Point of Care Using Isothermal DNA-Based Assays). ESKAPE is an abbreviation for their names and a reference to their capacity to get away from the impacts of generally utilized anti-infection agents through developmentally created systems.9‒13 Therapeutic herbs are more noteworthy to the soundness of individual and network.14 In this manuscript author exhibit different viewpoints foreseeing job of these mixes in grip of the bacterium to the host cell and accordingly accommodating in forestalling the disease and diminishes mortality because of this ailment.
Physiochemical properties
The physiochemical properties of the selected compounds were anticipated by openly accessible online SwissADME programming. Toxic capacity is imperative to assess in sedate planning as it help in deciding the harmful portion in animal model studies and decreases the quantity of animal model investigations.15,16
Molecular docking: receptor and ligand preparation
The 2D structures of Calotropis gigantea 20 phytoconstituents was downloaded by PubChem online server which is an openly accessible tool. The structures were then changed into. Mol2 design with the assistance of Chem3D Ultra, another bundle of Chem Office.17
Target selection and preparation
Prior detailed three-dimensional (3D) structure of the Crystal structure of S. aureus TyrRS in complex with SB-239629 (PDB ID: 1JIJ) was recovered from RCSB PDB.18 The water particles just as co-solidified ligands were removed from the PDB document and include polar hydrogen bonds.
Docking protocol
Before we moving to the molecular docking, the topological investigation of the protein structures including dynamic binding site/pocket was controlled by utilizing CASTp 3.0.19some residues are reasonable for anticipated ligand restricting site in the protein where the ligand can reversibly bind. Anyway other amino acid residues of the protein were giving right direction and affirmation.20 To decide the coupling mode and cooperation of the selected compounds and target, docking studies were performed by AutoDock/vina.21,22 The pdbqt files of the receptor protein, and C. gigantea compounds alongside the grid box getting at the dynamic site of the receptor for compound interaction was done through Auto Dock GUI program. The framework size limits along X, Y, and Z hub was scaled at 40 Å with a network dividing of 1 Å to permit appropriate binding adaptability at the docked site. The yield pdbqt documents were composed into a design (conf) file. The receptor was dealt with unbending element while ligands were kept adaptable to achieve the best fitting compliance regarding the receptor complex. The created arrangements of docking were grouped and those with root mean square deviation (RMSD) esteem <1.0 Å were viewed as just referred in given reference.23‒25 The coupling adaptation of ligands with the least binding affinity was portrayed as the steadiest compliance of the ligands as for the receptor.
Protein-ligand interaction studies
The interaction investigations of the protein-ligand complexed files perception were finished by PyMOL programming 1.3 variant. PyMOL can create large 3D picture of small atoms and protein. The polar (hydrogen bond) and non-polar connections among receptor and little ligand(s) were pictured by PyMOL programming. For binding investigation of the 2D cooperation, this was done by Discovery Studio 2019 (BIOVIA).26‒29
Calotropis gigantea flower extract phytoconstituent(s)
The phytoconstituent of C. gigantea flower extracts (Figure 1). The pharmacokinetics examination was done by SwissADME server where the atomic weight was denoted as a noteworthy quality of the in therapeutic method of the medication activity. In C. gigantea, natural compounds were in the range ≤500. Appropriately, RO5 the small atomic molecules of medications is transported, diffuse and ingested with no deterrent when contrasted with high sub-atomic weight. In compound examination RO5, subsequent stage is number of acknowledged hydrogen bond (O and N particles) and number of acceptor hydrogen bond (NH and OH) inside Lipinski's cut off points extend from 0-10 (H-bond acceptor) and 0-5 (H-bond contributor) separately. Lipophilicity (log P) and Topological Polar Surface Area (TSPA) values are critical properties for the gauge of oral risk of medication particles. The extending of log P from (0-5) or the majority of the mixes run from 0.94-5.00 (≤5), which is as far as possible for medication to infiltrate bio-layer. The computation of the surface regions which is involved by oxygen, nitrogen and through appended hydrogen iota is called TPSA. Along these lines, the TPSA is carefully related to the hydrogen holding capability of the compound. C. gigantea flower phytoconstituents were found to have the best medication like properties. For a medication, a decent bioavailability is more probable for mixes with ≤10 rotatable bonds and TPSA of ≤140å. All the TPSA, miLogP, number of particles, number of rotatable bonds and bioavailability (Table 1).
|
Compounds |
Formula |
MW |
RB |
HBA |
HBD |
MR |
logP |
RO5 |
|
2,3-Dihydro-benzofuran |
C8H8O |
120.15 |
0 |
1 |
0 |
35.79 |
1.89 |
Y |
|
2,4-dimethylacetophenone |
C10H12O |
148.20 |
1 |
1 |
0 |
46.57 |
2.11 |
Y |
|
2-Methoxy-4-vinylguaiacol |
C10H14O3 |
182.22 |
3 |
1 |
1 |
50.02 |
2.13 |
Y |
|
3,7-Dimethyl-1,6-octadien-3-ol |
C10H18O |
154.25 |
4 |
1 |
1 |
50.44 |
2.71 |
Y |
|
3,7-dimethyl-2,6-octadien-1-ol |
C10H18O |
154.25 |
4 |
1 |
1 |
50.40 |
2.75 |
Y |
|
3-Phenyl-2-propenoic acid |
C9H10N2O |
162.19 |
3 |
2 |
2 |
47.05 |
0.69 |
Y |
|
3-Thiophenemethanol |
C5H6OS |
114.17 |
1 |
1 |
1 |
30.45 |
1.54 |
Y |
|
5-Methyl-2,4-diisopropylphenol |
C13H20O |
192.30 |
2 |
1 |
1 |
62.59 |
2.86 |
Y |
|
6-hepten-1-ol, 2-Methyl |
C8H16O |
128.21 |
5 |
1 |
1 |
41.26 |
2.36 |
Y |
|
Benzaldehyde |
C7H6O |
106.12 |
1 |
1 |
0 |
31.83 |
1.36 |
Y |
|
Benzyl alcohol |
C7H8O |
108.14 |
1 |
1 |
1 |
32.57 |
1.66 |
Y |
|
Cinnamyl alcohol |
C9H10O |
134.18 |
2 |
1 |
1 |
42.50 |
1.98 |
Y |
|
Eugenol |
C10H12O2 |
164.20 |
3 |
2 |
1 |
49.06 |
2.37 |
Y |
|
Furfural |
C5H4O2 |
96.08 |
1 |
2 |
0 |
24.10 |
1.03 |
Y |
|
α-Terpineol |
C10H18O |
154.25 |
1 |
1 |
1 |
48.80 |
2.09 |
Y |
|
L-Tyrosine Methyl ester |
C10H13NO2 |
195.22 |
4 |
4 |
2 |
51.84 |
1.39 |
Y |
|
Phenethyl alcohol |
C8H10O |
122.16 |
2 |
1 |
1 |
37.38 |
1.70 |
Y |
|
Pinocampheol |
C10H18O |
154.25 |
0 |
1 |
1 |
46.86 |
2.34 |
Y |
|
Trans-3-Hexen-1-ol |
C6H12O |
100.16 |
3 |
1 |
1 |
31.64 |
1.94 |
Y |
Table 1 Physiochemical parameters of C. gigantea flower extract phytoconstituents
Docking studies
The prediction of the active binding site/pocket for S. aureus TyrRS (PDB ID: 1JIJ) was shown using CASTp 3.0 server (Figure 2). Molecular docking was done by AutoDock/vina21 using rigid docking method which was used because of the finding inhibitor and each step and binding must be examined. The grid box getting at the active site of the receptor for compounds binding was done through Auto Dock GUI program. Grid were generated, centre of the grid box X-11.179, Y-11.702 and Z-91.461 else the dimensions of the grid box X-20, Y-22 and Z-20 for the prepared proteins. For S. aureus TyrRS, the grid was generated around the bound ligand SB-239629 (2S)-2-[[(2S)-2-Amino-3-(4-hydroxyphenyl)propanoyl]amino]-2-[(2S,3S,4S,5S)-1,3,4,5-tetrahydroxy-4-(hydroxymethyl)piperidin-2-yl]acetic acid (Table 2). The docked PDB ID: 1JIJ bound ligand SB-239629 had docking score -7.5 kcal/mol and else the phytoconstituents of C. gigantea dock score against PDB ID: 1JIJ.
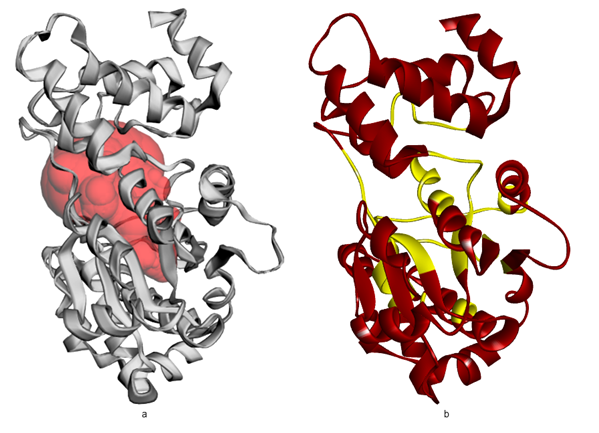
Figure 2 Cartoon representation of S. aureus TyrRS (A) the dot (red) representation in target protein showing active pocket by CASTp 3.0 (B) Crystal structure of S. aureus TyrRS (yellow) secondary structure visualize the region of amino acid involving active pocket by BIOVIA.
|
S.No. |
Compound |
Formula |
Docked score (kcal/mol) |
|
1 |
SB-239629 |
- |
-7.5 |
|
2 |
2,3-Dihydro-benzofuran |
C8H8O |
-5.8 |
|
3 |
2,4-dimethylacetophenone |
C10H12O |
-6.8 |
|
4 |
2-Methoxy-4-vinylguaiacol |
C10H14O3 |
-6.5 |
|
5 |
3,7-Dimethyl-1,6-octadien-3-ol |
C10H18O |
-5.2 |
|
6 |
3,7-dimethyl-2,6-octadien-1-ol |
C10H18O |
-5.5 |
|
7 |
3-Phenyl-2-propenoic_acid |
C9H10N2O |
-6.6 |
|
8 |
3-Thiophenemethanol |
C5H6OS |
-4.5 |
|
9 |
5-Methyl-2,4-diisopropylphenol |
C13H20O |
-6.5 |
|
10 |
6-hepten-1-ol, 2-Methyl |
C8H16O |
-4.9 |
|
11 |
Benzaldehyde |
C7H6O |
-5.2 |
|
12 |
Benzyl alcohol |
C7H8O |
-5.2 |
|
13 |
Cinnamyl alcohol |
C9H10O |
-5.7 |
|
14 |
Eugenol |
C10H12O2 |
-6.3 |
|
15 |
Furfural |
C5H4O2 |
-4.8 |
|
16 |
α-Terpineol |
C10H18O |
-6.5 |
|
17 |
L-Tyrosine_Methyl_ester |
C10H13NO2 |
-6.7 |
|
18 |
Phenethyl alcohol |
C8H10O |
-5.6 |
|
19 |
Pinocampheol |
C10H18O |
-5.1 |
|
20 |
Trans-3-Hexen-1-ol |
C6H12O |
-4.5 |
Table 2 Docking score for the phytoconstituents of C. gigantea flower extract with PDB ID: 1JIJ.
Protein-ligand complex interaction
After docking analysis the protein-ligand complex file analysing the interacting protein binding sites (amino acid), the phytoconstituents of C. gigantea flower extract against S. aureus tyrosyl-tRNA synthetase proteins (PDB:1JIJ) reveals, all the ligands binding active sites (means amino acid residues) occupy active pockets of the proteins (Figure 3). In these studies of ligand-receptor interactions depicts, S. aureus TyrRS target protein having highest interaction that is shown in (Figure 4). The ligand-receptor complex resolve the important conundrum for which residues are substantial for protein stabilization and otherwise important residue that are important for involved in protein conformation modification (Table 3).

PDB ID: 1JIJ_SB-239629.
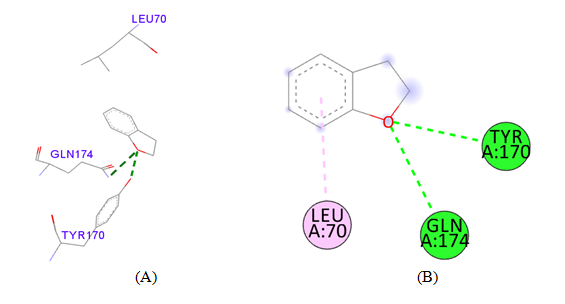
PDB ID: 1JIJ_2,3-Dihydro-benzofuran.
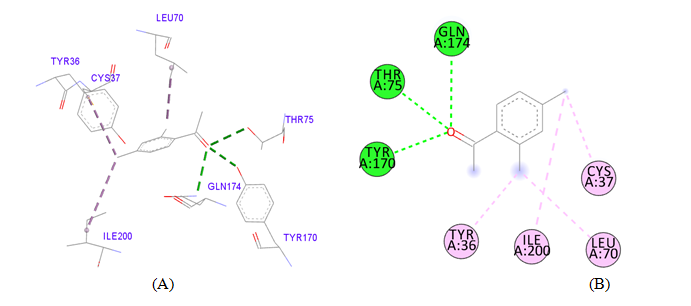
PDB ID: 1JIJ_2,4-dimethylacetophenone2,4-dimethylacetophenone.
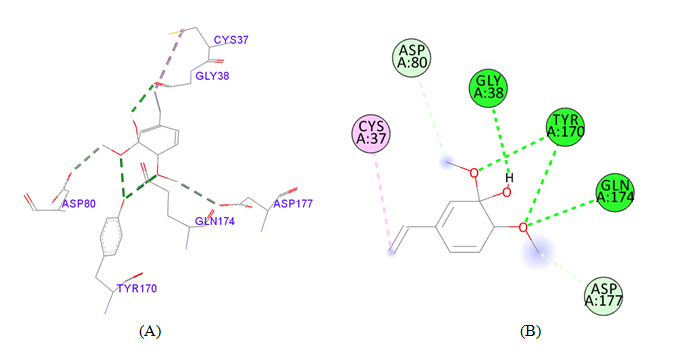
PDB ID: 1JIJ_2-Methoxy-4-vinylguaiacol.

PDB ID: 1JIJ_3,7-Dimethyl-1,6-octadien-3-ol.
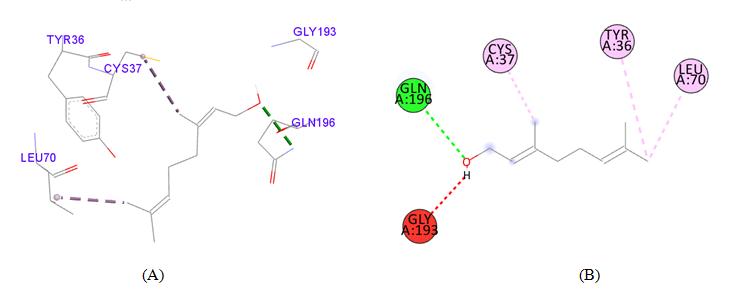
PDB ID: 1JIJ_3,7-dimethyl-2,6-octadien-1-ol.
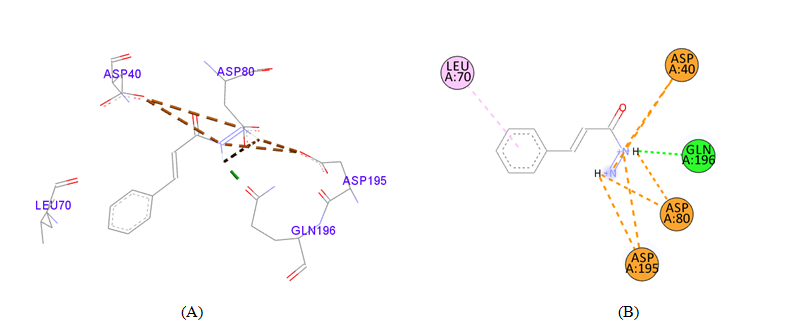
PDB ID: 1JIJ_3-Phenyl-2-propenoic acid.
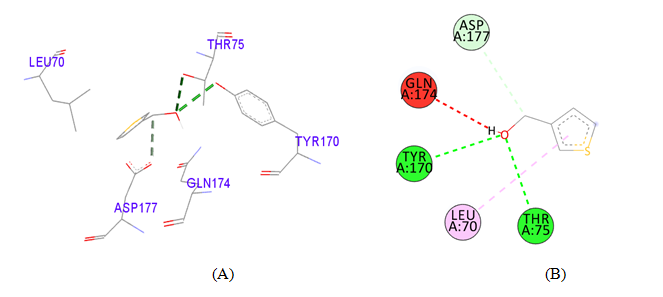
PDB ID: 1JIJ_3-Thiophenemethanol.
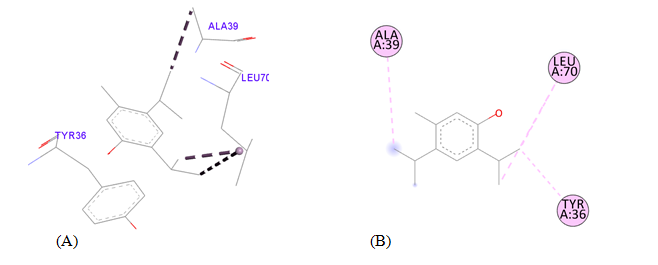
PDB ID: 1JIJ_5-Methyl-2,4-diisopropylphenol.
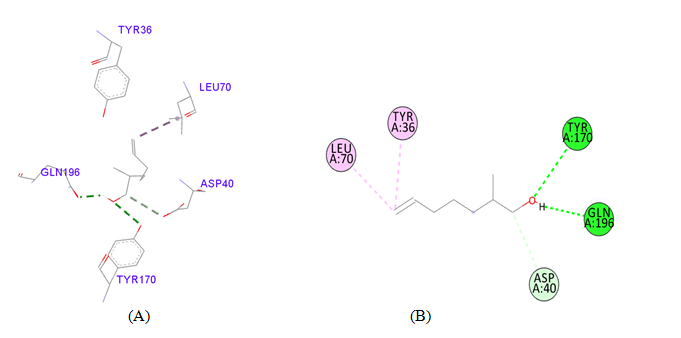
PDB ID: 1JIJ_6-hepten-1-ol, 2-Methyl.

PDB ID: 1JIJ_ Benzaldehyde.
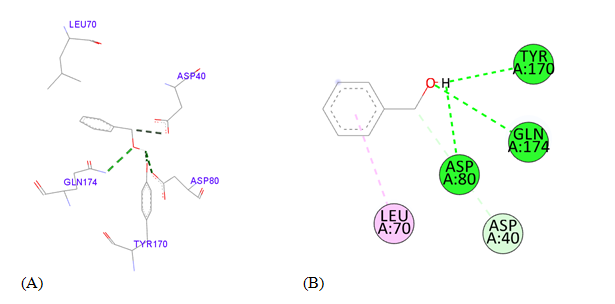
PDB ID: 1JIJ_ Benzyl alcohol.
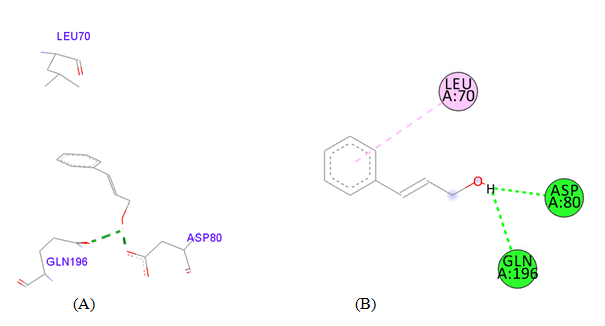
PDB ID: 1JIJ_ Cinnamyl alcohol.
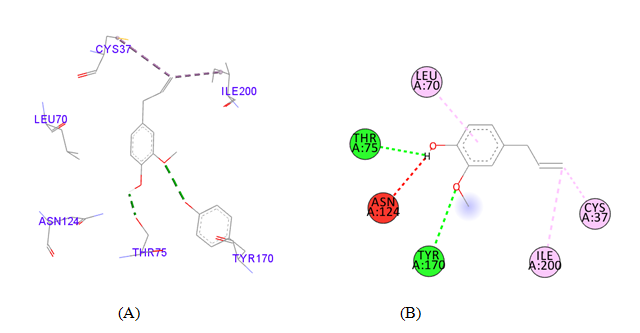
PDB ID: 1JIJ_Eugenio.
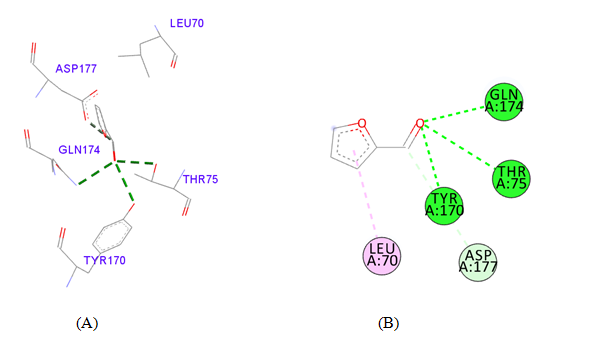
PDB ID: 1JIJ_Furfural.

PDB ID: 1JIJ_ α-Terpineol.
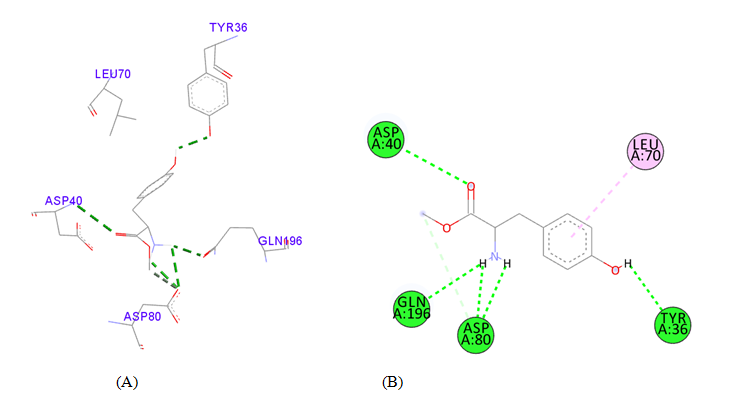
PDB ID: 1JIJ_ L-Tyrosine Methyl ester.

PDB ID: 1JIJ_ Phenethyl alcohol.

PDB ID: 1JIJ_ Pinocampheol.
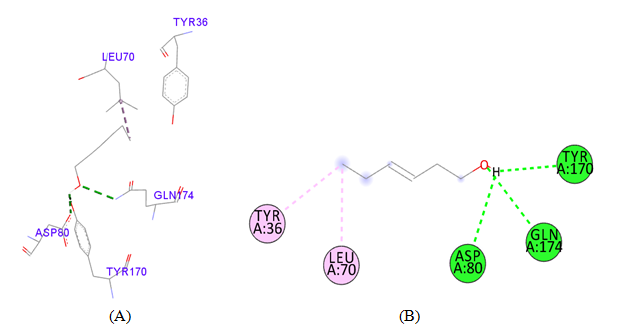
PDB ID: 1JIJ_ Trans-3-Hexen-1-ol.
Figure 3 Cartoon representation of S. aureus TyrRS (A) the dot (red) representation in target protein showing active pocket by CASTp 3.0 (B) Crystal structure of S. aureus TyrRS (yellow) secondary structure visualize the region of amino acid involving active pocket by BIOVIA.

Figure 4 Interaction studies of S. aureus TyrRS against natural compounds (A) Surface representation of the complex file showing deep cavity (B) Zoom image in the interactions of active site residues complex file (C) The interactions site showing the protein residues in S. aureus TyrRS protein. (D) Surface representation complex file showing the 2D interaction analysis done by Discovery studio (BIOVIA).
Ligand |
Interacting |
Distance (Å) |
Category |
Type |
SB-239629 |
Tyr36 |
3.08 |
H-Bond |
Conventional |
2,3-Dihydro-benzofuran |
Leu70 |
4.91 |
Hydrophobic |
Pi-Alkyl |
2,4-dimethylacetophenone |
Tyr36 |
5.02 |
Hydrophobic |
Pi-Alkyl |
2-Methoxy-4-vinylguaiacol |
Cys37 |
4.39 |
Hydrophobic |
Pi-Alkyl |
3,7-Dimethyl-1,6-octadien-3-ol |
Tyr36 |
4.98 |
Hydrophobic |
Pi-Alkyl |
3,7-dimethyl-2,6-octadien-1-ol |
Tyr36 |
5.25 |
Hydrophobic |
Pi-Alkyl |
3-Phenyl-2-propenoic_acid |
Asp40 |
4.72 |
H-Bond |
Salt Bridge |
3-Thiophenemethanol |
Leu70 |
5.30 |
Hydrophobic |
Pi-Alkyl |
5-Methyl-2,4-diisopropylphenol |
Tyr36 |
4.52 |
Hydrophobic |
Pi-Alkyl |
6-hepten-1-ol,2-Methyl |
Tyr36 |
4.78 |
Hydrophobic |
Pi-Alkyl |
Benzaldehyde |
Leu70 |
4.88 |
Hydrophobic |
Pi-Alkyl |
Benzyl alcohol |
Asp40 |
3.70 |
C H-Bond |
Conventional |
Cinnamyl alcohol |
Leu70 |
5.18 |
Hydrophobic |
Pi-Alkyl |
Eugenol |
Cys37 |
4.65 |
Hydrophobic |
Pi-Alkyl |
Furfural |
Leu70 |
4.86 |
Hydrophobic |
Pi-Alkyl |
α-Terpineol |
Cys37 |
4.48 |
Hydrophobic |
Pi-Alkyl |
L-Tyrosine_Methyl_ester |
Tyr36 |
2.30 |
H-Bond |
Conventional Pi-Alkyl |
Phenethyl alcohol |
Asp40 |
3.63 |
C H-Bond |
Conventional Pi-Alkyl |
Pinocampheol |
Asp40 |
2.16 |
Donor |
Unfavorable |
Trans-3-Hexen-1-ol |
Tyr36 |
5.26 |
Hydrophobic |
Pi-Alkyl |
Table 3 Detailed molecular interactions obtained following the rigid ligand docking of PDB ID: 1JIJ
gigantea flower extract naturals compounds, the physiochemical analysis and RO5 analysis shows that all those compound having >500 MW, >10 number of rotatable bonds (RB), hydrogen bond donor and acceptor >5 bonds and logP value is >3 all the properties showing these compounds are good for bioavailability of the drug.6‒8,14,15 The predicted ligand binding site for S. aureus TyrRS protein showing pocket residues 36-54, 70-91, 170-196 and 221-241.18,19 Using docking analysis, the natural compounds against S. aureus TyrRS having top 5 docking scores compounds are 2,4-dimethylacetophenone (-6.8), L-Tyrosine_Methyl_ester (-6.7), 3-Phenyl-2-propenoic_acid (-6.6), 2-Methoxy-4-vinylguaiacol (-6.5) and α-Terpineol (-6.5).20,21,23 Protein-ligands interaction studies of S. aureus TyrRS ternary complexes with natural compounds as the putative inhibitor. In complex file interaction studies showing all these natural compounds closely interacted with active sites /substrate binding domains. The interaction residues of the ternary structure of S. aureus TyrRS protein are Tyr36, Cys37, Gly38, Ala39, As40, Leu70, Asp80, Thr75, Tyr170, Gln174, Asp177, Gly193, Asp195, Gln196, Ile 200. This interaction study confirmed that there was the involvement of Hydrogen bond (H-bond), Carbon-Hydrogen bond (C-H bond) and Pi-Alkyl bond. The interaction study showing the strong interaction between S. aureus TyrRS protein with natural compounds but the L-Tyrosine_Methyl_ester docking showing the maximum four hydrogen bond interaction with TYR36, ASP40, ASP80 and GLN19 residues and al the residues are closely interacted with active sites.26‒29 The outcome was determined that the phytocompounds of C. gigantea can be used as a natural better therapeutic target for antimicrobial resource and therefore targeting natural compounds might be an advantageous step in the direction of therapeutic development.
A C. gigantea flower part with auxiliary highlights of flavonoids content was read for their antibacterial and cancer prevention agent profiles. The C. gigantea flower progressive has most noteworthy antibacterial action against an ESKAPE pathogenic strains of Gram-negative and Gram-positive microbes, including a MRSA strain, just as a fascinating cancer prevention agent profile. The occurrence of a phytoconstituents could be significant as it is an auxiliary component of the most dynamic substances. These substances or compounds could be considered for its antibacterial potential and could be a significant hotspot for the plan and advancement of new foreign agent of infective material. As the most dynamic compound for additional examinations, docking concentrates with significant objective were performed to explain a possible system of activity for this compound. PDB ID:1JIJ, tyrosyl-tRNA synthetase from S. aureus were studied as potential targets, and a correlation between the observed inhibitory activity and the in silico molecular docking scores was obtained. Moreover, compounds also approved by RO5 drug likeness properties. Future studies will be carried out to evaluate the anti-biofilm activity of this compound and to explore the biological activities of the metal complexes to find new compounds with increased antimicrobial potential and a broader spectrum activity.
The authors acknowledge support from the Centre for Interdisciplinary Research in Basic Science, Jamia Millia Islamia University. Md. Amjad Beg also acknowledges University Grants Commission Maulana Azad National Fellowship for the financial support and Jamia Millia Islamia University.
The authors declare that they have no potential conflict of interests.

©2020 Amjad, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.