eISSN: 2379-6367


Research Article Volume 8 Issue 3
Centre for Interdisciplinary Research in Basic Science, Jamia Millia Islamia, India
Correspondence: Dr Fareeda Athar, Associate Professor, Centre for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia, New Delhi-110025, India, Tel +91-11-26981717, Fax +91-11.26980164
Received: May 28, 2020 | Published: June 15, 2020
Citation: Beg MA, Athar F. Anti-HIV and Anti-HCV drugs are the putative inhibitors of RNA-dependent-RNA polymerase activity of NSP12 of the SARS CoV2 (COVID-19). Pharm Pharmacol Int J. 2020;8(3):163-172. DOI: 10.15406/ppij.2020.08.00292
Corona virus is the emerging infectious viral agent that had to threaten the world with its epidemic since 2003 with SARS and 2013 with MERS virus. This virus again stuck the whole world with its robust pandemic in 2019whena novel form of the corona virus was emerged in and infect the local population of Wuhan, China. The virus named as SARS CoV-2 and the disease caused by this infectious virus is introduced as COVID 19. Since December 2019, this virus took more than 432, 973 lives all around the world. This study elaborates the role of HIV and HCV drug in targeting nsp12 gene of SARS CoV 2 that is responsible for RNA dependent RNA polymerase activity. The work employed homology modelling, structure validation and molecular docking analysis to evaluate the binding affinity and interaction analysis between NSP12 (RdRp) protein and HIV/HCV drugs. Outcome of the study impacted on Nelfinavir (NFV), Raltegravir (RAL) and Delavirdine (DLV) among Anti-HIV and Paritaprevir (PTV), Beclabuvir (BCV) and Ledipasvir (LDV) among Anti-HCV as the most effective inhibitors of SARS CoV-2 RdRp ternary complexes. The drugs have a strong binding affinity with the residues that are present in the active site of RdRp of the virus and essential for its replication. This study establishes significant information in the direction of therapeutic development as we are dealing with the situation where we urgently required medication or vaccine to combat COVID-19. Therefore, this study provides essential molecular information about the FDA approved antiviral drugs that can be used to treat this disease. Importantly, the listed drugs had never prioritized for their effectiveness against COVID-19.
Keywords: COVID-19, NSP12 (RdRp), antiviral drug, molecular dockingv
NCBI, National Centre for Biotechnology Information; SARS CoV-2, severe acute respiratory syndrome Corona virus 2; RdRp, RNA dependent RNA polymerase; CoVs, Coronaviruses; NSP, non-structural protein; NSPs, non-structural protein sequences; PDB, protein data bank; 3D, three dimensional; FDA, Food and Drugs Administration; HCV, Hepatitis C virus; HCMV, human cytomegalovirus; HSV, herpes simplex virus; HPV, human papillomavirus; HIV, human immunodeficiency virus; VZV, Varicella zoster virus; +ssRNA, single stranded RNA of positive sense
The start of the year 2020 also creates the epidemic situation in the world as a series of confounded influenza-like cases were reported in China in December 2019. In a very short period, this epidemic changed into a dangerous pandemic and spread to other countries. WHO reported that this pandemic is due to novel Corona virus (2019-CoV) which is a new virus in corona virus group that cause COVID-19.1-3 The virus can invade into the respiratory, gastrointestinal, hepatic, and central nervous system of human, livestock, birds, bat, mouse, and many other wild animals. This COVID-19 outbreak in china had broken all the parameters and measures that occur in 2002 due to severe acute respiratory syndrome (SARS) in 2002/2003 and the Middle East respiratory syndrome (MERS).4,5 It is enormously conceivable that novel CoVs emergence is inescapable in the future due to changes in the genome, climate and ecology, and the increased interactions of human with animals. These viruses are one of the largest virus group of order Nidovirales. Nidovirales consists of Coronaviridae, Arteriviridae, Mesoniviridae and Roniviridae families. Coronaviridae family is further subdivided into Coronavirinae and Toronavirinae. Coronavirinae is comprised of four genera i.e. alpha, beta, gamma and delta. Novel corona virus or SARS CoV-2 is a part of Beta corona virus genera. SARS CoV-2 is closely related to two relatively bat corona viruses known as bat-SL-CoVZC45 and bat-SL-CoVZXC21 with almost 88% similarity. This similarity range is much different from the SARS corona virus (79%) and MERS corona virus (50%). Moreover, one of the recent reports suggests pangolins as a possible virus reservoir. Pangolin-CoV is 91.02% and 90.55% identical to SARS-CoV-2 and one bat corona virus RaTG13 respectively and the S1 protein of CoV is very much identical to the virus samples obtained from dead pangolin lung samples.6 Although there have been multiple reports are present on the human transmission of the virus, the exact virus reservoir is remained to find.
The genome is single-stranded RNA of positive sense (+ssRNA) of approximately 30kb size.7,8 There are mainly two types of proteins that are characterized by structural and non-structural proteins. Structural proteins include spikes, nucleocapsids, matrix and envelope proteins whereas non-structural proteins (NSPs) comprise of RNA dependent RNA polymerase protein (RdRp) that is coded by NSP12 gene.9,10 The first ORFs covers almost two-thirds of the whole genome length that encodes 16 NSPs. Ribosomal frame shifting between ORF1a and ORF1b leads to the production of polyproteins 1a (pp1a) and polyprotein 1b (pp1b) respectively. RdRp is an essential enzyme for running life cycle of various RNA viruses and therefore continuously been targeted to eradicate RNA viruses. The genome of CoVs contains 6 ORFs, envelope protein, membrane glycoprotein, nucleocapsid phosphoprotein and surface protein. RdRp is a crucial enzyme in the life cycle of RNA viruses, including coronaviruses. RdRp is targeted in different RNA viruses, including Hepatitis C Virus (HCV), Zika Virus (ZIKV), and coronaviruses (CoVs).11-16
Suddenly this disease grabs the attention of all due to a tremendous increase in the number of incidences and deaths cases in China. Till 26th May 2020, total 5,631,824 COVID-19 confirmed cases, 2,399,258 recovered cases and 348,965 deaths were reported in around 205 countries areas and territories.17 The infrequent arrival and epidemics prompt us that this CoVs is an unembellished universal threat to health. Therefore, there is an imperative prerequisite to developed advanced and effective therapies and vaccines against CoVs. The current study provides some essential aspects about NSP12 gene of SARS CoV-2 that codes RNA dependent RNA polymerase enzyme (RdRp). The enzyme is a pronounced target for previous therapeutic approaches to target several RNA viruses. RdRp is a conserved protein in the corona virus family and comprise two conserved aspartate residues in the active site of the enzyme. Controlling RNA virus infection is a critical task because these viruses mutate with greater velocity as compare to any DNA virus. The study includes homology modelling of NSP12 gene using template-based modelling, its validation and molecular docking analysis with Human Immuno Deficiency Virus (HIV) drugs, HCV drugs and anti-malarial drugs.18,19
Homology modelling of SARS CoV-2RdRp
Complete genome sequence of SARS CoV-2 is available in the National Centre for Biotechnology Information (NCBI) with the accession no. (NC_045512.2) was used for homology modelling.20 The homology modelling was done by using the SWISS-MODEL server which is freely available, and it built the protein model with the given target-template sequence alignment.20 The identity matrix showed that NSP12 protein from SARS-CoV is 97% identical with the SARS CoV-2 (orf1ab gene sequence). The identical sequence template PDB ID: 6NUR confirmed which is 97.05% identical to NSP12.17 The predicted 3D model structure validation was done by SAVES meta server of the University of California Los Angles (UCLA) where some server RAMPAGE, Verify3D and ERRAT which predicted different stereochemical properties.21-23 The final validated model was qualified for protein quality prediction by using ProQ server which was then optimized to find correct models in contrast to other methods. The protein quality prediction measured two types of predicted score LGscore and MaxSub. LGscore is -log of a P-value and MaxSub ranges from 0-1, where 0 is insignificant and 1 most significant.24-26 Before moving to the molecular docking, the topological properties of protein structures, including binding pockets was determined by using CASTp 3.0. Few residues are responsible for predicted ligand binding site in the protein where the ligand can reversibly bind. However other amino acid residues of the protein were providing correct orientation and confirmation.27,28
Molecular docking
Preparation of Ligand and Receptor
Docking studies were accomplished on Acer Laptop with Intel® Core i3 8th Gen, 12 GB RAM and 1 TB hard disk running on Windows 10 operating system. Antiviral compounds which wereFood and Drugs Administration (FDA) approved drugs proposed from the literature and were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/) in the SDF format.29 Chem 3D Pro 14.0 was used for making the 3D structure of the antiviral drugs which is enlisted in Supplementary Table S1.30 The downloaded files were converted by the PyRx 0.8 tool (FastSpring, Amsterdam, Netherlands) were minimize the 3D structure and produce the dock file in PDBQT format. AutoDockVina is an open-source program which was utilized in all docking experiments.31,32 74 compounds were tested against SARS CoV-2 RdRp where 70 compounds were antiviral drugs against different viruses like (HCV, HCMV, HIV, HSV, HPV, VZV and Influenza viruses) and rest compounds are in a clinical trial or already published as effective in various manner.
|
S. No |
Drug name |
Abbreviation |
Virus Family |
|
1 |
Abacavir |
ABC |
HIV |
|
2 |
Acyclovir |
ACV |
HSV, VZV |
|
3 |
Adefovir |
- |
HBV |
|
4 |
Amantadine |
AMT |
Influenza virus A |
|
5 |
Amprenavir |
APV |
HIV-1 |
|
6 |
Asunaprevir |
ASV |
HCV-1 |
|
7 |
Atazanavir |
ATV |
HIV |
|
8 |
Beclabuvir |
BCV |
HCV-1 |
|
9 |
Boceprevir |
BOC |
HCV-1 |
|
10 |
Brivudine |
BVDU |
HSV-1, VZV |
|
11 |
Catechin |
SINE |
HPV |
|
12 |
Cidofovir |
CDV |
HCMV |
|
13 |
Cobicistat |
COBI |
HIV |
|
14 |
Daclatasvir |
DCV |
HCV-1 |
|
15 |
Darunavir |
DRV |
HIV |
|
16 |
Dasabuvir |
DAS |
HCV-1 |
|
17 |
Delavirdine |
DLV |
HIV-1 |
|
18 |
Didanosine |
ddI |
HIV |
|
19 |
Dolutegravir |
DTG |
HIV |
|
20 |
Docosanol |
C22 |
HSV |
|
21 |
Efavirenz |
EFV |
HIV-1 |
|
22 |
Elbasvir |
EBR |
HCV-1 or 4 |
|
23 |
Elvitegravir |
EVG |
HIV |
|
24 |
Emtricitabine |
(-) FTC |
HIV |
|
25 |
Entecavir |
ETV |
HBV |
|
26 |
Entecavirine |
ETR |
HIV-1 |
|
27 |
Famciclovir |
FCV |
HSV, VZV |
|
28 |
Favipiravir |
FPV |
Influenza virus A, B & C |
|
29 |
Fosamprenavir |
FPA |
HIV-1 |
|
30 |
Foscarnet |
PFA |
HCMV, HSB |
|
31 |
Ganciclovir |
GCV |
HCMV |
|
32 |
Grazoprevir |
GZR |
HCV-4 |
|
33 |
Idoxuridine |
IDU |
HSV-1 |
|
34 |
Indinavir |
IDV |
HIV |
|
35 |
Lamivudine |
3TC |
HIV, HBV |
|
36 |
Laninamivir |
- |
Influenza virus A &B |
|
37 |
Ledipasvir |
LDV |
HCV-1 |
|
38 |
Letermovir |
- |
HCMV |
|
39 |
Lopinavir |
LPV |
HIV |
|
40 |
Maraviroc |
MVC |
HIV |
|
41 |
Nelfinavir |
NFV |
HIV |
|
42 |
Nevirapine |
NVP |
HIV-1 |
|
43 |
Ombitasvir |
OBV |
HCV-1 |
|
44 |
Oseltamivir |
OTV |
Influenza virus A & B |
|
45 |
Paritaprevir |
PTV |
HCV-1 |
|
46 |
Penciclovir |
PCV |
HSV |
|
47 |
Peramivir |
PRV |
Influenza virus A & B |
|
48 |
Podofilox |
PDX |
HPV |
|
49 |
Raltegravir |
RAL |
HIV |
|
50 |
Ribavirin |
RBV |
HCV-1 |
|
51 |
Rilpivirine |
RPV |
HIV-1 |
|
52 |
Rimantadine |
RIM |
Influenza virus A |
|
53 |
Ritonavir |
RTV |
HIV |
|
54 |
Saquinavir |
SQV |
HIV |
|
55 |
Simeprevir |
SMV |
HCV-1 |
|
56 |
Sofosbuvir |
SOF |
HCV-3 |
|
57 |
Stavudine |
d4T |
HIV |
|
58 |
Telaprevir |
TVR |
HCV-1 |
|
59 |
Telbivudine |
LdT |
HBV |
|
60 |
Tenofovir alafenamide |
TAF |
HIV |
|
61 |
Tenofovir |
- |
HIV |
|
62 |
Tipranavir |
TPV |
HIV-1 |
|
63 |
Trifluridine |
TFT |
HSV |
|
64 |
Valacyclovir |
VACV |
HSV, VZV |
|
65 |
Valganciclovir |
VGCV |
HCMV |
|
66 |
Vaniprevir |
VPV |
HC-1 |
|
67 |
Vidarabine |
VDR |
HSV, VZV |
|
68 |
Zalcitabine |
ddC |
HIV |
|
69 |
Zanamivir |
ZAN |
Influenza virus A & B |
|
70 |
Zidovudine |
AZT |
HIV |
Table S1 Antiviral drugs against SARS COV-2 NSP 12 (RdRp)
Top hit selection against SARS CoV-2 RdRp
Molecular docking analysis of SARS HCoV-2 RdRp was done by determining binding affinity scores (docking scores) based on top 10 hits were selected out of 74 antiviral drugs. The selected drugs further analyzed based on the interactions where false-positive were avoided and highly interactive partners were selected having a binding pocket of SARS CoV-2 RdRp.
Interaction analysis: Visualization and Evaluation
The interaction analysis of the protein-ligand and interaction of the binding sites were studied by using PyMOL and Chimera.33-35 The possible dock conformations of the 2D ligand-receptor interactions was accomplished using Discovery Studio.36 Further, binding analysis using the visualization approach was carried out to understand the binding pattern of the drug with protein.37-41
Modelling of SARS CoV-2 RdRp
Homology modelling of SARS CoV-2 RdRp was done which showed the sequence identity 97.05% with the template PDB ID: 6NUR (SARS-NSP12) (Figure 1). This 6NUR structure is the complex structure of NSP12 with NSP7 and NSP8 with 3.1 Å resolution.17 The generated SARS CoV-2 RdRp structure QMEAN (qualitative model energy analysis) score is -0.68 which was the optimized score for further study.20 The model also includes two zinc ions and it encompasses residues 4501-5315 of the SARS CoV-2 orf1ab. The model was validated by SAVES, the Verify3D server evaluated the scoring as 91.16% of the residues have average 3D-1D score >= 0.2 and the score pass while the quality factor predicted by ERRAT server was 95.9839.22,23 The Ramachandran plot showed that 92.5% residues were in the allowed region, 7.2% residues in the additional allowed region 0.3% else in the generously allowed region and no residues in the disallowed region (see the Ramachandran plot in Figure S1).21 The protein quality estimation by ProQ server found the LG score was 6.636 and MaxSub score was 0.610 which was predicted to be asquality of good model.26 Superimposed cartoon model of SARS-NSP12 and SARS CoV-2 RdRp in red active sites residues region (Figure 2a & 2b) was shown using CASTp 3.0 serve. The SARS CoV-2 RdRp model showed the A chain and two cofactor ions between triphosphate moiety.27
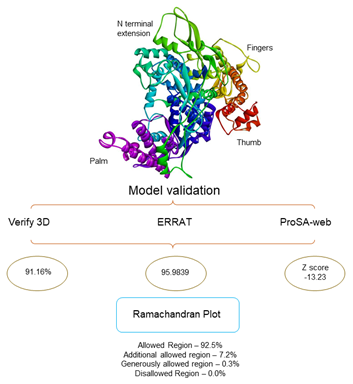
Figure 1 Homology modelling and Model validation: The SARS CoV-2 RdRp model built by Swiss model server with template PDBID: 6NUR and sequence identity is 97.05%. Model validation by SAVES metaserver were stereochemistry of the model was good, with 100% residues are in allowed region.
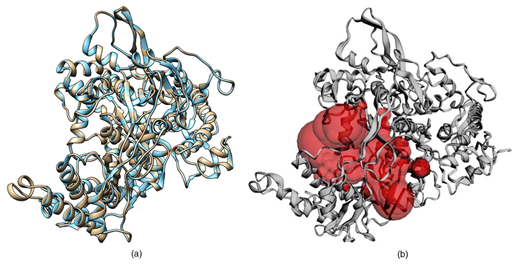
Figure 2 The three-dimensional structure SARS HCoV-2 RpRd: (a) The superimposed three-dimensional cartoon representation of SARS CoV-2 RpRd in (Grey) and the SARS-CoV RdRp in (Sky blue) by using Chimera. (b) Homology model of SARS CoV-2 RpRd predicted binding site residues in (red color) using CASTp 3.0.
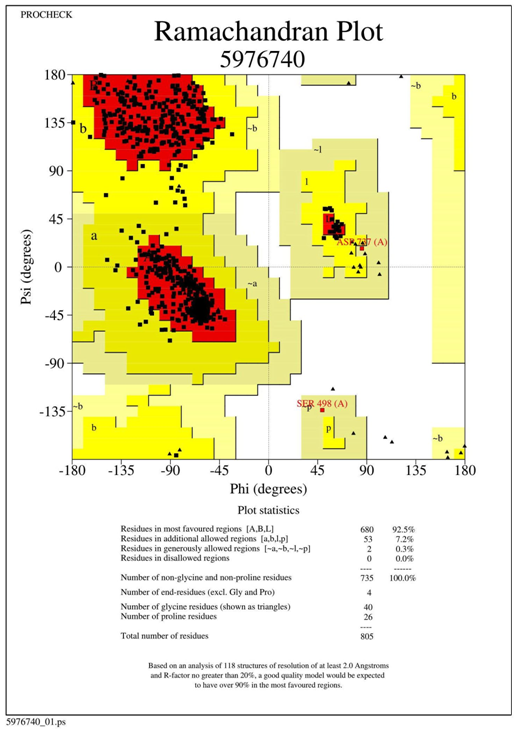
Figure S1 Ramachandran plot by RAMPAGE: In Ramachandran plot shows 100% residues are in allowed region.
Docking identifies (+ssRNA) antiviral drugs as a potent inhibitor
In molecular docking, the screening of 74 antiviral drugs produced log files, where the affinity (kcal/mol) obtained and docked poses for discrete compounds were analyzed. This log-file screen-out because of the analysis of binding score or docking score and the docking scores are enlisted in Supplementary Table S2. Molecular docking result having best binding affinity (docking score) was screened based on all the log files and output files and top 19 antiviral drugs were selected that were screened based on binding energies (kcal/mol) (Figure 3).32,41 HIV drugs (Delavirdine, Dolutegravir, Indinavir, Maraviroc, Nelfinavir, Raltegravir, Rilpivirine, Ritonavir, Saquinavir and Tipranavir) and HCV drugs (Beclabuvir, Boceprevir, Daclatasvir, Elbasvir, Ledipasvir, Paritaprevir, Simeprevir, Sofosbuvir and Vaniprevir) was used in the study were enlisted in Table 1.29 Furthermore, the interaction analysis of selected drugs with the SARS CoV-2 RdRp on the specific sites highlights that the selected antiviral drug putatively acts as an inhibitor against the respective enzymes.
|
S.No. |
Drugs Name |
Molecular formula |
Molecular weight |
Docking score |
|
1 |
Abacavir |
C14H18N6O |
286.33 |
-7.1 |
|
2 |
Acyclovir |
C8H11N5O3 |
225.2 |
-5.9 |
|
3 |
Adefovir |
C8H12N5O4P |
273.19 |
-6.7 |
|
4 |
Amantadine |
C10H17N |
151.25 |
-5.5 |
|
5 |
Amprenavir |
C25H35N3O6S |
505.6 |
-7.2 |
|
6 |
Asunaprevir |
C35H46ClN5O9S |
748.3 |
-7.9 |
|
7 |
Atazanavir |
C38H52N6O7 |
704.9 |
-7.5 |
|
8 |
Azithromycin |
C38H72N2O12 |
749 |
-7.9 |
|
9 |
Beclabuvir |
C36H45N5O5S |
659.8 |
-9.9 |
|
10 |
Boceprevir |
C27H45N5O5 |
519.7 |
-8 |
|
11 |
Brivudine |
C11H13BrN2O5 |
333.13 |
-6.2 |
|
12 |
Catechin |
C15H14O6 |
290.27 |
-7.5 |
|
13 |
Cidofovir |
C8H14N3O6P |
279.19 |
-5.7 |
|
14 |
Cobicistat |
C40H53N7O5S2 |
776 |
-7.8 |
|
15 |
Daclatasvir |
C40H50N8O6 |
738.9 |
-8.4 |
|
16 |
Darunavir |
C27H37N3O7S |
547.7 |
-7.9 |
|
17 |
Dasabuvir |
C26H27N3O5S |
493.6 |
-8.8 |
|
18 |
Delavirdine |
C22H28N6O3S |
456.6 |
-8.5 |
|
19 |
Didanosine |
C10H12N4O3 |
236.23 |
-6.2 |
|
20 |
Dolutegravir |
C20H19F2N3O5 |
419.4 |
-8 |
|
21 |
Efavirenz |
C14H9ClF3NO2 |
315.67 |
-7.7 |
|
22 |
Elbasvir |
C49H55N9O7 |
882 |
-9.5 |
|
23 |
Elvitegravir |
C23H23ClFNO5 |
447.9 |
-7.3 |
|
24 |
Emtricitabine |
C8H10FN3O3S |
247.25 |
-6 |
|
25 |
Entecavir |
C12H15N5O3 |
277.28 |
-7.1 |
|
26 |
Etravirine |
C20H15BrN6O |
435.3 |
-7.1 |
|
27 |
Famciclovir |
C14H19N5O4 |
321.33 |
-6.3 |
|
28 |
Favipiravir |
C5H4FN3O2 |
157.1 |
-5.3 |
|
29 |
Fosamprenavir |
C25H36N3O9PS |
585.6 |
-6.9 |
|
30 |
Foscarnet |
CH3O5P |
126.01 |
-4.8 |
|
31 |
FV 100 |
C27H35N3O6 |
497.6 |
-7.8 |
|
32 |
Ganciclovir |
C9H13N5O4 |
255.23 |
-6.9 |
|
33 |
Grazoprevir |
C38H50N6O9S |
766.9 |
ND |
|
34 |
Hydroxychloroquine |
C18H26ClN3O |
335.9 |
-6 |
|
35 |
Idoxuridine |
C9H11IN2O5 |
354.1 |
-6 |
|
36 |
IDX-184 |
C25H35N6O9PS |
626.6 |
-7.3 |
|
37 |
Imiquimod |
C14H16N4 |
240.3 |
-6.8 |
|
38 |
Indinavir |
C36H47N5O4 |
613.8 |
-8.4 |
|
39 |
Lamivudine |
C8H11N3O3S |
229.26 |
-5.6 |
|
40 |
Laninamivir |
C13H22N4O7 |
346.34 |
-7 |
|
41 |
Ledipasvir |
C49H54F2N8O6 |
889 |
-9.7 |
|
42 |
Letermovir |
C29H28F4N4O4 |
572.5 |
-8.3 |
|
43 |
Lopinavir |
C37H48N4O5 |
628.8 |
-7.9 |
|
44 |
Maraviroc |
C29H41F2N5O |
513.7 |
-8.1 |
|
45 |
Nelfinavir |
C32H45N3O4S |
567.8 |
-8.8 |
|
46 |
Nevirapine |
C15H14N4O |
266.3 |
-7.1 |
|
47 |
Ombitasvir |
C50H67N7O8 |
894.1 |
-7.8 |
|
48 |
Oseltamivir |
C16H28N2O4 |
312.4 |
-5.6 |
|
49 |
Paritaprevir |
C40H43N7O7S |
765.9 |
-10 |
|
50 |
Penciclovir |
C10H15N5O3 |
253.26 |
-5.4 |
|
51 |
Peramivir |
C15H28N4O4 |
328.41 |
-7.3 |
|
52 |
Podofilox |
C22H22O8 |
414.4 |
-7.3 |
|
53 |
Raltegravir |
C20H21FN6O5 |
444.4 |
-8.7 |
|
54 |
Ribavirin |
C8H12N4O5 |
244.2 |
-6.3 |
|
55 |
Rilpivirine |
C22H18N6 |
366.4 |
-8.4 |
|
56 |
Rimantadine |
C12H21N |
179.3 |
-6.1 |
|
57 |
Ritonavir |
C37H48N6O5S2 |
720.9 |
-8 |
|
58 |
Saquinavir |
C38H50N6O5 |
670.8 |
-9.6 |
|
59 |
Simeprevir |
C38H47N5O7S2 |
749.9 |
-9.4 |
|
60 |
Sofosbuvir |
C22H29FN3O9P |
529.5 |
-8 |
|
61 |
Stavudine |
C10H12N2O4 |
224.21 |
-6.1 |
|
62 |
Telaprevir |
C36H53N7O6 |
679.8 |
-7.9 |
|
63 |
Telbivudine |
C10H14N2O5 |
242.23 |
-6.9 |
|
64 |
Tenofovir alafenamide |
C21H29N6O5P |
476.5 |
-7.1 |
|
65 |
Tenofovir |
C9H14N5O4P |
287.21 |
-6.1 |
|
66 |
Tipranavir |
C31H33F3N2O5S |
602.7 |
-8.2 |
|
67 |
Trifluridine |
C10H11F3N2O5 |
296.2 |
-6.7 |
|
68 |
Valacyclovir |
C13H20N6O4 |
324.34 |
-6.2 |
|
69 |
Valganciclovir |
C14H22N6O5 |
354.36 |
-6.6 |
|
70 |
Vaniprevir |
C38H55N5O9S |
757.9 |
-9.9 |
|
71 |
Vidarabine |
C10H13N5O4 |
267.24 |
-6.4 |
|
72 |
Zalcitabine |
C9H13N3O3 |
211.22 |
-5.8 |
|
73 |
Zanamivir |
C12H20N4O7 |
332.31 |
-6.4 |
|
74 |
Zidovudine |
C10H13N5O4 |
267.24 |
-6.3 |
Table S2 Docking Score (binding affinity) of antiviral drugs against SARS COV-2 NSP 12 (RdRp)
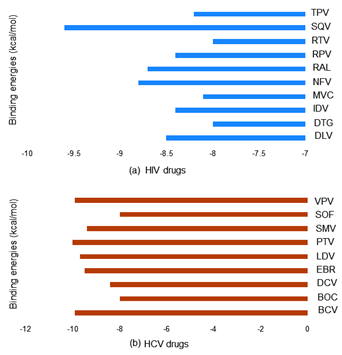
Figure 3 Top hit selected by molecular docking: By using AutoDock Vina molecular docking experiment calculated top binding energies (docking scores) of HIV drugs in (blue) are Tipranavir (TPV), Saquinavir (SQV), Ritonavir (RTV), Rilpivirine (RPV), Raltegravir (RAL), Nelfinavir (NFV), Maraviroc (MVC), Indinavir (IDV), Dolutegravir (DTG), Delavirdine (DLV) and HCV drugs in (brown) are Vaniprevir (VPV), Sofosbuvir (SOF), Simeprevir (SMV), Paritaprevir (PTV), Ledipasvir (LDV), Elbasvir (EBR), Daclatasvir (DCV), Boceprevir (BOC), Beclabuvir (BCV) against SARS CoV-2 RdRp.
|
S.No. |
Drug name |
Abv. |
Docking score |
Mode of action |
|
HIV Drugs |
||||
|
1 |
Delavirdine |
DLV |
-8.5 |
High toxicity not to be used as much |
|
2 |
Dolutegravir |
DTG |
-8 |
Targets HIV integrase to inhibit the integration of viral DNA into human chromosomes |
|
3 |
Indinavir |
IDV |
-8.4 |
Blocks the active site of HIV protease to prevent cleavage of viral precursor proteins |
|
4 |
Maraviroc |
MVC |
-8.1 |
Blocks GP120-CCR5 interaction to inhibit HIV entry |
|
5 |
Nelfinavir |
NFV |
-8.8 |
Blocks the active site of HIV protease to prevent cleavage of viral precursor proteins |
|
6 |
Raltegravir |
RAL |
-8.7 |
Targets HIV integrase to inhibit the integration of viral DNA into human chromosomes |
|
7 |
Rilpivirine |
RPV |
-8.4 |
Binds directly to HIV RT and inhibits DNA synthesis |
|
8 |
Ritonavir |
RTV |
-8 |
Blocks the active site of HIV protease to prevent cleavage of viral precursor proteins |
|
9 |
Saquinavir |
SQV |
-9.6 |
Blocks the active site of HIV protease to prevent cleavage of viral precursor proteins |
|
10 |
Tipranavir |
TPV |
-8.2 |
Blocks the active site of HIV protease to prevent cleavage of viral precursor proteins |
|
HCV drugs |
||||
|
1 |
Beclabuvir |
BCV |
-9.9 |
Inhibits activity of the VZV DNA polymerase |
|
2 |
Boceprevir |
BOC |
-8 |
Protease inhibitor |
|
3 |
Daclatasvir |
DCV |
-8.4 |
Targets NS5A and NS3/4A protease to prevent HCV replication |
|
4 |
Elbasvir |
EBR |
-9.5 |
Inhibit activities of NS5A and NS3/4A protease, respectively |
|
5 |
Ledipasvir |
LDV |
-9.7 |
Harvoni inhibits HCV NS5A and NS5B polymerase to prevent RNA replication |
|
6 |
Paritaprevir |
PTV |
-10 |
Inhibits activities of HCV NS5A, NS5B polymerase, and NS3/4A protease |
|
7 |
Simeprevir |
SMV |
-9.4 |
Target HCV NS5B and NS3/4 protease, respectively |
|
8 |
Sofosbuvir |
SOF |
-8 |
Target HCV NS5B and NS3/4 protease, respectively |
|
9 |
Vaniprevir |
VPV |
-9.9 |
Protease inhibitor |
Table 1 Selected antiviral drugs against SARS COV-2 NSP 12 (RdRp)
Interaction studies of antiviral drugs against SARS CoV-2 RdRp
The interaction analysis of SARS CoV-2 RdRp ternary complexes suggested the in HIV-drugs as the putative inhibitor against SARS CoV-2 RdRp isNelfinavir (NFV), Raltegravir (RAL) and Delavirdine (DLV) shown in Figure 4 and in HCV-drugs suggested inhibitor Paritaprevir (PTV), Beclabuvir (BCV) and Ledipasvir (LDV) shown in Figure 5 have closely interacted with active sites and substrate binding domains.33 The interaction study suggested that the active sites consist of several important active site residues involved in the interaction are VAL301, LYS302, ASN434, ARG346, LYS391, SER392, ASN398, LYS402, LYS436, THR437, ARG444, VAL448, ARG460, LEU467, ASP509, CYS513, ASP514, ARG515, SER570, THR571, SER573, ALA576, THR578, ALA579, ASN582, SER650, ASP651 and ASP736. This interaction study confirmed that there was the involvement of Carbon-Hydrogen bond (C-H bond), Hydrogen bond (H-bond), Pi-Anion bond (π-Anion), Pi- Sigma bond (π-Sigma), Amide Pi-stacked bond (Amide π stacked) and Alkyl bond. The important points of interaction study between antiviral drugs against SARS CoV-2 RdRp were enlisted in Table 2.36 The interaction study also suggested that the interaction between the drug and RdRp enzyme is a strong interaction and therefore targeting this enzyme with the listed inhibitors might be an advantageous step in the direction of therapeutic development.
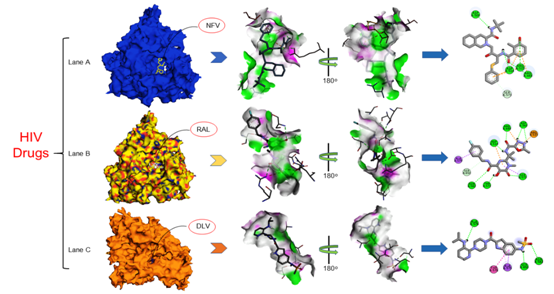
Figure 4> Interaction studies of SARS CoV-2 RdRp against HIV drugs Nelfinavir (NFV), Raltegravir (RAL), Delavirdine (DLV) drugs: Crystal structure of SARS CoV-2in (Lane A) Interactions of active site residues in SCoV 2-NFVcomplex (Lane B) Interactions of active site residues in SCoV 2-RALcomplex (Lane C) Interactions of active site residues in SCoV 2-DLVcomplex and the 2D interaction analysis done by Discovery studio.
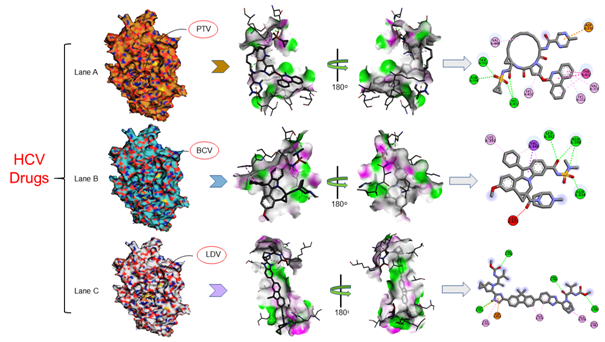
Figure 5 Interaction studies of SARS CoV-2 RdRp against HCV drugs Paritaprevir (PTV), Beclabuvir (BCV), Ledipasvir (LDV) drugs: Crystal structure of SARS CoV-2in (Lane A) Interactions of active site residues in SCoV 2-PTVcomplex (Lane B) Interactions of active site residues in SCoV 2-BCVcomplex (Lane C) Interactions of active site residues in SCoV 2-LDVcomplex and the 2D interaction analysis done by Discovery studio.
|
S.No. |
Compound Name |
Interacting Residues |
|||||
|
C-H bond |
H-bond |
π-Anion |
π-Sigma |
Amide π stacked |
Alkyl |
||
|
HIV Drugs |
|||||||
|
1 |
Delavirdine |
ALA 576 |
ALA 579 |
THR 578 |
|||
|
ASN 582 |
|||||||
|
SER 650 |
|||||||
|
2 |
Nelfinavir |
SER 573 |
ARG 444 |
||||
|
ASP 509 |
|||||||
|
ASP 514 |
|||||||
|
ARG 515 |
|||||||
|
3 |
Raltegravir |
SER 650 |
ASP 343 |
ARG 515 |
|||
|
ARG 444 |
|||||||
|
ASP 514 |
|||||||
|
THR 571 |
|||||||
|
SER 573 |
|||||||
|
ASN 582 |
|||||||
|
HCV Drugs |
|||||||
|
1 |
Beclabuvir |
SER 392 |
VAL 448 |
LYS 391 |
|||
|
ASN 398 |
|||||||
|
ASN 434 |
|||||||
|
2 |
Ledipasvir |
LYS 391 |
ASP 651 |
LEU 467 |
|||
|
ARG 446 |
CYS 513 |
||||||
|
ARG 460 |
ALA 576 |
||||||
|
ASP 514 |
ALA 579 |
||||||
|
3 |
Paritaprevir |
SER 392 |
ASP736 |
THR 437 |
VAL 301 |
||
|
ASN 398 |
LYS302 |
||||||
|
LYS 402 |
LYS 436 |
||||||
|
|
|
|
|
|
|
|
VAL 448 |
Table 2 Selected antiviral drugs with their interacting residues of SARS COV-2 NSP 12 (RdRp)
We are currently dealing with the global health emergency of COVID-19 pandemic. The disease almost covers every country with devastating nature in China, Italy, United States of America, Brazil and Spain. The disease had already accounted for almost 432, 973 deaths on the worldwide level. It is the third major outbreak of corona virus after SARS and MERS epidemic. Antiviral drugs have been developing using that target viral RNA polymerase. The present study deals with anti-viral drugs to declare their effectiveness against COVID-19. The study includes Anti-HIV drugs Delavirdine, Nelfinavir and Raltegravir and Anti-HCV drugs Beclabuvir, Ledipasvir and Paritaprevir most effective binding partner among various listed drugs. The binding study establishes a significant platform for further testing the binding parameters in in-vitro condition and beneficial for determining inhibitory effect of the inhibitor against the enzyme. Especially, the advanced therapeutic target generation would need essential clinical and experimental studies for further and strongly confirming drug effectiveness against disease.
The authors acknowledge support from the Centre for Interdisciplinary Research in Basic Science, Jamia Millia Islamia University. Md. Amjad Beg also acknowledges University Grants Commission Maulana Azad National Fellowship for the financial support and Jamia Millia Islamia University.
The authors declare that they have no potential conflict of interests.
Md Amjad Beg: Conceptualization idea, Methodology designing, Software handling, writing manuscript and evaluate results.
Dr Fareeda Athar: Reviewing and corresponding author.

©2020 Beg, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.