MOJ
eISSN: 2381-182X


Research Article Volume 2 Issue 6
1Institute of Food Research, UK
2Department of Chemical Sciences, University of Naples Federico II, Italy
3Department of Agriculture, University of Naples ?Federico II?, Italy
4School of Medicine, The University of Manchester, UK
Correspondence: Concetta Valeria L Giosafto, Department of Chemical Sciences, University of Naples Federico II Parco Gussone, 80055 Portici, Naples, Italy, Tel 0039 081 2539470
Received: June 30, 2016 | Published: September 2, 2016
Citation: Giosafto CVL, Rigby N, Sorrentino A, et al. Optimization of in vitro n-deglycosylation of ovomucoid protein. MOJ Food Process Technol. 2016;2(6):205-212. DOI: 10.15406/mojfpt.2016.02.00058
Glycosylation is a common post-translational modification that confers important properties to proteins. Glycans are implicated in a wide range of intracellular, cell-cell and cell-matrix recognition events and, therefore, are of great biological interest. In this paper we have taken an enzymatic approach by using peptide N-glycosidase F to release N-linked glycans from ovomucoid. Our study revealed that the protein in its native conformation was susceptible to this enzyme. Nevertheless, denaturation by means of both heat (at 100°C) and reducing agent treatment, before the enzymatic exposure, greatly enhanced the extent of deglycosylation.
SDS-PAGE and Western blot analyses revealed that the intact protein migrates as a diffuse band that was attributed to glycosylation heterogeneity. The complete deglycosylation of the protein resulted in a shift of the protein migration and in the formation of a sharp protein band and it was confirmed by gel filtration, FTIR and MALDI-TOF analyses. Importantly, LC-MS/MS revealed for the first time the presence of eight potential deglycosylation sites on the protein.
Keywords: enzymatic deglycosylation, pngase F, protein denaturation, maldi-TOF, LC-MS/MS
Glycoproteins have a wide distribution in nature and serve a vast number of functions. Of all the biologically occurring macromolecules, the glycoproteins, which consist of carbohydrate moieties covalently linked to a polypeptide backbone, represent the most diverse group, ranging from substances in which the carbohydrate component represents less than 1% of the total weight to those in which it represents over 80% of the total.1 Glycosylation is a common posttranscriptional modification of proteins. Covalently bound sugar residues stabilize proteins, act as recognition sites, and provide loci for immuno response.2 Carbohydrate modifications of proteins fall into three general categories: N-linked modification of asparagine (Asn), O-linked modification of serine or threonine and glycosylphosphatidyl inositol derivatization of the C-terminus carboxyl group.3 Each of these transformations is catalyzed by one or more enzymes which demonstrate different peptide sequence requirements and reaction specificities. In particular, N-linked glycosylation is catalyzed by a single enzyme, oligosaccharyl transferase (OT), and involves the co-translational transfer of a lipid-linked tetradecasaccharide (GIcNAca-Mang-Glc3) to an Asn side chain within a nascent polypeptide.4,5
The ovomucoid protein, the dominant allergen of egg white,6 possesses prominent carbohydrate domains. In fact, it contains as much as 25% carbohydrates, present as oligosaccharides, each joined to the polypeptide chain by an asparagyl residue.7-9 Ovomucoid has a molecular weight of 28 kDa and an isoelectric point of 4.1, and comprises 11% of egg white proteins.7 It is composed of 186 aminoacids, arranged in three tandem domains, each about 60 amino acids in length, called Kazal like-domains, because they are homologous to the pancreatic secretory trypsin inhibitor (Kazal inhibitor).8 Each domain is cross linked by three intra-domain disulfide bonds; however, they lack any inter-domain disulfide bonds.10 It is a very stable protein since it is not denatured by trichloric acid-acetate procedures or 8 M urea.11,12 The carbohydrate chains are penta-antennary, heterogeneous and partially sialylated, resulting in substantial mass ad charge heterogeneity of native ovomucoid. Various chemical and enzymatic methods are possible for the removal of glycans from glycoproteins. Peptide-N4-(acetyl-β-glucosaminyl) asparagines amidase (PNGase F) is one of the most widely used enzymes for the removal of the N-linked glycans from glycoproteins.13-15 This enzyme has a broad substrate specificity, hydrolyzes at the glycosylamine linkage and generates a carbohydrate-free peptide and an intact oligosaccharide with the di-N-acetylchitobiose unit at the reducing agent.16 This enzymatic release of N-glycans is also accompanied by the deamidation of Asn residue to aspartic acid which provides an indirect indication of N-glycosylation sites of a glycoprotein.17,18
This article examines the enzymatic in vitro deglycosylation of ovomucoid using PNGase F. The efficiency of both heat and reducing agent on the extent of deglycosylation was evaluated. Deglycosylation of the protein was assessed on the basis of the shift in molecular weight associated with the sugar removal by SDS-PAGE and Western blot. The absence of carbohydrate from ovomucoid was also established by different analytical techniques such as Infrared Spectroscopy (FTIR), Gel Filtration (GF), Matrix Assisted Laser Desorption Ionisation Time-of-Flight Mass Spectrometry (Maldi-TOF-MS) and by Liquid Chromatography coupled with tandem mass spectrometry (LC-MS/MS).
Materials
Partially purified ovomucoid (Product T9253; trypsin inhibitor III-O: chicken egg white) and the Protein Deglycosylation Kit (Product code PP0201) were purchased from Sigma-Aldrich, (Dorset, UK). A rabbit polyclonal antibody anti-ovomucoid was purchased from InCura srl (Cremona, Italy). Goat anti-rabbit IgG (whole molecule) alkaline phosphatase conjugate was obtained from Sigma-Aldrich, (Dorset, UK). Gels, reagents and protocols for analytical SDS-PAGE were from Invitrogen, Renfrewshire, UK and for immunoblotting were from Bio-Rad (Hertfordshire, UK). All other chemicals were of AR grade unless specified and were obtained from Sigma-Aldrich, unless specified.
Methods
Purification of ovomucoid: Ovomucoid purification was performed by anion exchange chromatography on Q Sepharose FF (XK 16/20, GE Healthcare) in 20 mM Triethanolamine (TEA), pH 7.3 and eluted with a linear gradient from 0 to 100% of 1 M NaCl. Selected fractions were dialyzed against H2O (Medicell International, England, molecular weight cut off 10-14 kDa membrane) and submitted to Size Exclusion Chromatography (SEC) on Superdex S- 200 prep grade (Pharmacia; 1.6 × 60 cm) in 25 mM Tris 150 mM NaCl pH 7.5 (1 mL min-1). The eluent was monitored for protein by following the absorbance at 280 and 220 nm. Selected fractions were again dialyzed against water before being freeze dried and stored at -20 °C until required.
Protein determination: Protein determination was carried out by the Coomassie Plus Protein Assay Reagent (Thermo Scientific, Rockford, USA), using bovine serum albumin as standard.19
Deglycosylation: Deglycosylation of ovomucoid was performed according with technical instructions obtained from the Protein Deglycosylation Kit with some modifications. Deglycosylation was based on the enzymatic reaction using either 2.5 U or 5 U of PNGase F. Before the enzyme addition, a denaturing solution (containing 2% octyl β-D-glucopyranoside with 100 mM 2-mercaptoethanol) and/or heating treatment (100 °C for 10 min, 30 min and 60 min) were used for denaturing the protein. In some cases one of the two treatments or both were omitted in order to study their effectiveness on the extent of deglycosylation. The control sample was carried out by incubating the protein under the same conditions in the absence of PNGase F.
Circular dichroism (CD) and Fourier transformed infrared spectroscopy (FTIR): Protein secondary structure was determined using a combination of CD and FTIR. Both native and denatured ovomucoid was examined. In particular, for studying the effect of heating and reducing agent on the secondary structure of the molecule, the protein was pre-treated for 60 min at 100 °C and incubated in the presence of 30 mM dithiothreitol (DTT) (60 min at 37 °C) and 60 mM iodoacetamide (60 min at 37 °C), dialyzed over night against H20 by using small dialysis cups (Slide-A-Lyzer MINI Dialysis Units, Thermo Scientific, molecular weight cut-off 10 kDa) and then analyzed. Far UV CD spectra of purified native ovomucoid were recorded with a JASCO J-180 spectropolarimeter (Jasco, Essex, UK) at 20 °C in aqueous solutions. 50 μL protein samples (1 μg μL-1) were measured in quartz cuvettes of 0.1 mm path length. All spectra were baseline corrected using the corresponding aqueous solution. Absorption between 180 and 260 nm was monitored at 0.5 nm intervals. The spectra obtained represent an average of four consecutive scans and the mean residue ellipticity was expressed as deg cm2 dmol-2.
FTIR spectra were measured with a FTS 175C spectrometer (Bio-Rad, US) equipped with a MCT detector and a single-reflection diamond ATR sampling accessory (GoldenGate, Specac). 10 μL of 1 μg μL-1 protein solution, previously dialyzed against water (10-14 kDa membrane), were dried down with a stream of dry air and the spectra were measured. 256 scans at 2 cm-1 resolution were averaged and referenced against the empty crystal. To aid structural interpretation, the spectra were Fourier deconvoluted with the spectrometer software, using a K factor of 1.7, and a half-width of 17 cm-1. Fourier deconvolution was not carried out for analyzing the differences between intact and deglycosylated ovomucoid because of the low signal intensity.
SDS-PAGE (Sodium Dodecyl Sulfate-PolyAcrylamide Gel Electrophoresis) analysis: SDS-PAGE was run in NuPage 4-12% Bis-Tris precast gels from Invitrogen in MES buffer according to manufacturer’s instructions. Samples were reduced by using Reducing agent (containing 500 mM dithiothreitol) (Invitrogen), dissolved in lithium dodecyl sulphate sample buffer 4× (Invitrogen) and heated for 10 min at 70 °C prior to analysis. Gels were fixed in 40% v/v methanol, 10% v/v acetic acid and after 30 min were rinsed in deionised water prior to staining with Colloidal Blue Staining Kit (Invitrogen). SeeBlue Plus2 Pre-Stained Standards (Invitrogen) were run in each gel.
Western blot: Western blotting was performed using nitrocellulose (NC) membranes after SDS-PAGE according to described method [20]. Briefly, semi-dry blotting of proteins from gels to NC membrane (0.2 μm pore, Trans-Blot transfer media, Bio-Rad) in 39 mM glycine, 48 mM Tris base, 0.0375% SDS, 20% methanol, pH 8.8–8.9 was carried out using a Trans-Blot SD, semi-dry transfer cell (Bio-Rad) at 15 V for 20 min. Membranes were blocked by incubation (1 h, 20 °C) in 5% skimmed milk powder (Marvel, Premier Foods, Lincolnshire, UK) in PBST (137 mM NaCl, 0.01 M Na phosphate, 2.7 mM KCl, 0.05% v/v Tween 20, pH 7.4) followed by washing in PBST (3×5 min, 50 mL). The membrane was incubated using gentle shaking in a 1:5000 dilution of anti-ovomucoid for 1 h at 18 °C, then washed in PBST (as above). The blot was then incubated with gentle shaking in a 1:5000 dilution of goat anti-rabbit alkaline phosphatase conjugate for 2 h at 20 °C and then washed in PBST (as above). Bound antibody was located by briefly washing the membrane in water (1×30 s, 100 mL) and then staining using 10 mL of a SIGMA FASTTM BCIP/ NBT substrate tablet solution (1 tablet in 10 mL of water).
Analytical Gel Filtration (GF): 1 mg of the intact and deglycosylated ovomucoid forms was analyzed by size exclusion chromatography on a Superdex 75 gel permeation column (10 × 300 mm; GE Healthcare), equilibrated and eluted in 25 mM Phosphate buffer pH 7.0, 150 mM NaCl, at flow rate 0.4 mL min-1. The absorbance was monitored at 280 and 220 nm. The columns were calibrated with a set of gel filtration molecular weight standards (Bio-Rad, US). The molecular size standards (Bio-Rad Laboratories, Hercules, Calif.) (mass in kDa; Stokes radius in Å) were bovine thyroglobulin (670; 85), bovine gammaglobulin (158; 52.5), chicken ovalbumin (44; 30.5), horse myoglobin (17; 19), and vitamin B12 (1.357 kDa; 7.5); The Stokes radii and the apparent molecular masses of the eluting proteins were estimated by fitting the corresponding elution volume data to, respectively, the plots of the Stokes radius and molecular mass value vs elution volume for the standard proteins.
Analysis of protein intact mass by Matrix-Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS): Analysis of intact protein mass was performed by mixing each sample with a saturated sinapinic acid (Sigma-Aldrich, Dorset, UK) matrix in 30% (v/v) acetonitrile, 0.1% (v/v) trifluroacetic acid (TFA). The target plates used were polished stainless steel (Bruker Daltonics, Coventry, UK). Approximately 0.5 μL of sample/matrix mixture was spotted onto the MALDI target and dried in air. The MALDI-MS measurements were performed using a Bruker UltraFlex MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Coventry, UK) equipped with a pulsed N2 laser (λ = 337 nm, frequency 10 Hz). Whole protein spectra were recorded over the 4000-40000 m/z range in linear mode at an accelerating voltage of 25 kV by averaging of 300 individual laser shots. The concentration of each protein solution was adjusted to give peak intensity similar to that of the calibrants used. Calibration was performed using ubiquitin and myoglobin.
Characterization of ovomucoid peptides by MS/MS
In Gel Digestion: In Gel digestion was performed according to Rigby et al.20 Briefly, gel plugs were first conditioned with two 15-min incubations in 200 mM ammonium bicarbonate (ABC) in 50% acetonitrile (Solution B) followed by a 10-min incubation with acetonitrile (200 µL) before air drying. Cysteine residues were then reduced by incubation with 10 mM DTT in 50 mM ABC (200 µL) for 30 min at 60 °C before being alkylated with 100mM iodoacetamide in 50 mM ABC (200 µL) for 30 min at room temperature. The gel plugs were then reconditioned with a further two 15 min incubations in Solution B (200 µL) followed by 10 min in acetonitrile (200 µL) before air drying. Digestion at 37 °C for 3 hours used either 50 ng (5µL /well) porcine trypsin (sequencing-grade, Promega) dissolved in 10 mM ABC or 50 ng (5µL/well) Chymotrypsin (sequencing grade, Roche) dissolved in 20 mM ABC containing 10 mM CaCl2. Digestion was stopped and peptides were extracted with 5% formic acid (5 µL each tube).
The supernatant from the in-gel digest was recovered from digest reaction tube and put into a 1.5 ml reaction tube. To extract further peptides 20 µl 50% Acetonitrile was added to the gel piece and left at room temperature for 3-5 mins before the liquid removed and added to the Eppendorf tube. The sample was then dried down at the Low Drying setting (no heat) on a Speed Vac SC110 (Savant) fitted with a Refrigerated Condensation Trap and a Vac V-500 (Buchi). The sample was frozen at -70 °C or immediately reconstituted for LC-MS/MS analysis.
To prepare the sample for the LC-MS/MS analysis the sample was redissolved in 30-100 µl 0.5% formic acid and vortexed for 20 seconds before sitting in a sonicating water bath (Kerry) for 5 minutes at room temperature to facilitate dissolution. The sample was then briefly pulse centrifuged before the appropriate volume (25-50 µl) was dispensed into a 0.2 ml skirted 96-well PCR plate (ThermoAB-800). Once all samples were added to the plate it was sealed with adhesive PCR foil seal.
Liquid chromatography–mass spectrometry/mass spectrometry (LC-MS/MS) analysis: LC-MS/MS analysis was performed using a LTQ-Orbitrap mass spectrometer (Thermo Electron) and a nanoflow-HPLC system (nanoACQUITY: Waters). Peptides were trapped on line to a Symmetry C18 Trap (5 µm, 180 µm x 20 mm) which was then switched in-line to a UPLC BEH C18 Column, (1.7 µm, 75 µm x 250 mm) held at 45 °C. Peptides were eluted by a gradient of 0-80% acetonitrile in 0.1% formic acid over 50 min at a flow rate of 250 nL min−1. The mass spectrometer was operated in positive ion mode with a nano-spray source at a capillary temperature of 200 °C.
The Orbitrap was run with a resolution of 60 000 over the mass range m/z 300–2000 and an MS target of 106 and 1 s maximum scan time. The MS/MS was triggered by a minimal signal of 2000 with an Automatic Gain Control target of 30000 ions and maximum scan time of 150 ms. For MS/MS events selection of 2+ and 3+ charge states selection were used. Dynamic exclusion was set to 1 count and 30 s exclusion time with an exclusion mass window of ±20 ppm.
Identification of N-Glycosylation sites using Mascot: Ovomucoid sequences identified from the mfg file (prepared from the Orbitrap-derived RAW data file using DTA supercharger) using an in–house version of the Mascot LC-MS-MS search tool from Matrix Science. The search parameters were as follows: the taxonomy group searched was Chordata in the SPtrEMBL database, tryptic digest was assumed to have a maximum of up to two missed cleavages, the carbamidomethylation of cysteine residues was a fixed modification and oxidation of methionine residues was a variable modification, peptide masses were monoisotopic and either 2+ or 3+ charged, the peptide and fragment mass tolerances were set at 10 ppm and 0.5 Da respectively. To search for deglycosylation sites a variable modification for the deamidation of Asn was also used. To identify any unexpected modifications, or non-specific cleavages an error tolerant search was also performed. Individual ions scores >36 (for Trypsin) and >40 (for Chymotrypsin) indicated identity or extensive homology (p-value <0.05).
FTIR analysis
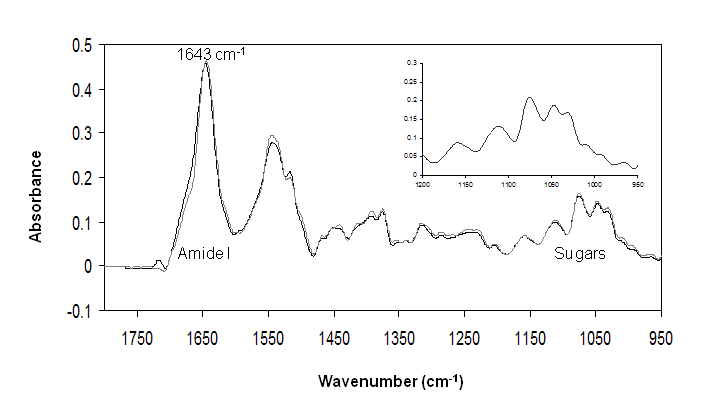
The absorption spectra of ovomucoid untreated and treated for 60 min at 100 °C is reported in Figure 1 over the range 1800-950 cm-1. According to literature,21 the peak at 1643 cm-1 of Amide I is assigned to random coil structures. The unordered conformation (usually referred to as random coil) is, in fact, usually associated with the IR band between 1640 and 1648 cm-122.
The high intensity in the region 1200-900 cm−1, corresponding to the carbohydrate band can be also observed in the spectrum. This spectrum shows the large contribution of carbohydrates to ovomucoid molecule. It is possible to note that the heat treatment does not lead to changes in the protein secondary structure and no obvious changes in glycosylation to ovomucoid (Figure 1), indicating the heat stability of the protein. Moreover, the shape of peaks in the region 900-1200 cm-1 resembles those of N-acetylglucosamine (GlcNAc) as indicated by Khajehpour et al.2
CD analysis
It was found necessary to denature the protein prior to deglycosylation because the steric hindrance imparted by the oligosaccharides limited access of the enzyme.16 In the following sections we will show that only limited deglycosylation of the native ovomucoid was possible with PNGase F.
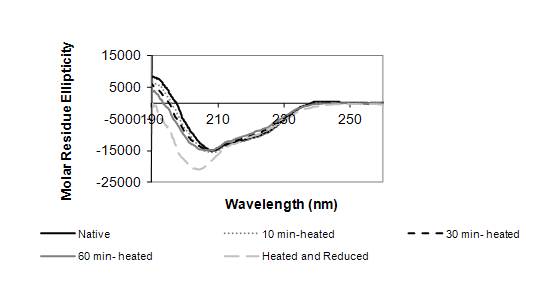
The change in conformation of ovomucoid following both the heat treatment and denaturing agents was monitored by CD analysis (Figure 2). The CD spectroscopy analysis identified the native protein with a folded structure, containing α-helices, β-sheets and prominently unordered structures. As shown in Figure 2, the heat treatment alone did not significantly change the secondary structure of ovomucoid, revealing the thermal stability of the protein. In contrast, both the incubation for 60 min at 100 °C and the treatment with the reducing agents showed a significantly increased negative dichroism with a minimum at 208 nm. This indicates a change to less ordered structures.
Analysis of ovomucoid deglycosylated forms
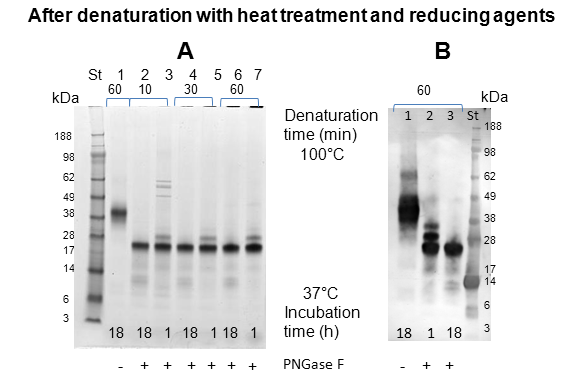
SDS-PAGE of intact and deglycosylated ovomucoid: One of the simplest methods to assess the extent of deglycosylation is by mobility shift on SDS-PAGE. The deglycosylation reaction was carried out following either heat-treatment of the protein at 100 °C for different times (10 min, 30 min, 60 min), or by using denaturing agents β-mercaptoethanol and octyl-glycoside. Subsequently, the protein was incubated either for 1 h or 18 h with PNGase F. At the end of the incubation the reaction products were analyzed by SDS-PAGE (4-12%) (Figure 3 & 4). Initially, the band corresponding to intact ovomucoid appeared quite broad owing to the heterogeneity of the oligosaccharides, as shown in the lanes 1 of Figure 3, with an apparent molecular weight (Mr) of ~ 40 kDa. Figure 3 shows that in all cases the enzymatic reaction using PNGase F resulted in a similar extent of deglycosylation, in fact the deglycosylation reaction converted the ~40 kDa band to a ladder of faster migrating polypeptides (Figure 3, Panel A-C) suggesting that the protein under these conditions is only partially deglycosylated even after 18 h incubation with the enzyme. It is worth pointing out that ovomucoid was not fully deglycosylated in its native form even when incubated with excess of PNGase F for 18 h (5 U instead of 2.5 U) (Figure 3, lane 4, Panel A). Therefore, the same experiments were also carried out by denaturing the protein both with heat treatment and the addition of reducing agent (Figure 4). Figure 4, Panel A demonstrates that these treatments together increased the susceptibility of the protein to deglycosylation. SDS-PAGE analysis (Figure 4, Panel A, lane 1) revealed again that the intact protein migrated as a diffuse band. However, the denaturation of protein prior the deglycosylation resulted in a significant shift of the protein migration, independent of the incubation time at 100 °C. In fact, when the protein was incubated for 18 h with PNGase F, the ladder of polypeptides, shown in Figure 3, was converted to a sharp single band with a molecular weight of a Mr of ~ 20 kDa (Figure 4, Panel A, lanes 2,4,6), whereas when the incubation was performed for only 1 h, an additional band of ~ 22 kDa was also detected (lanes 3, 5, 7). This behaviour was also confirmed by Western blot experiment (Figure 4, Panel B), where the broad and diffuse appearance of the intact protein band was even more evident (Figure 4, Panel B, lane 1). The Western blot shows that, even under denaturing conditions, the deglycosylation carried out for 1 h was only partial, whereas ovomucoid incubated for 18 h with the PNGase F was observed as a single band of a Mr of ~ 20 kDa.
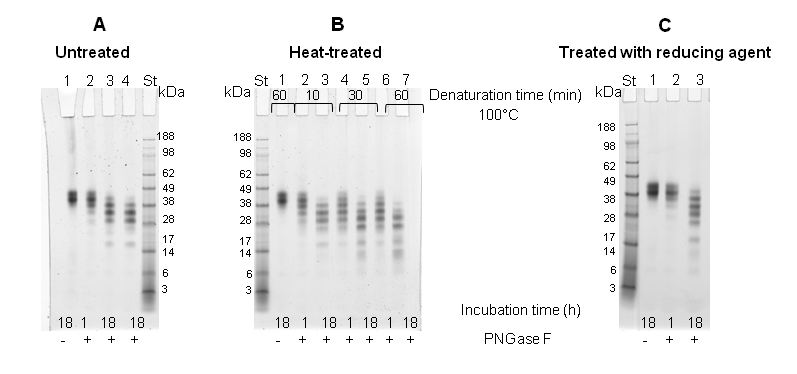
Ovomucoid was thought to contain 5 potential N-linked glycosylation sites.8 The attached oligosaccharides are of a high-mannose type and correspond to 25 % of the molecular weight. The molecular weight of the protein is 28 kDa,7 although the apparent molecular weight, according to SDS-PAGE, is approximately 35-38 kDa (Figure 3 & 4, lanes 1). Thus it should decrease by about 7 kDa after complete deglycosylation. The results shown in Figure 4 correspond to these values.
The sample incubated in the presence of PNGase F for 18 h previously denatured by heat treatment (60 min at 100 °C) and by using reducing agent, in which the full deglycosylation was observed in SDS-PAGE (Figure 4, Panel A, lane 6) was further analyzed by FTIR, Gel filtration, MALDI-TOF and Orbitrap. The control, represented by the protein treated under the same conditions but in the absence of deglycosidase, was also analyzed (Figure 4, Panel A, lane 1).
GF: The elution behaviour of the intact and deglycosylated protein was analyzed by analytical GF in order to observe the differences in size between the two forms. Distinct peaks with different retention properties for the intact and deglycosylated ovomucoid were observed (Figure 5, Panel A), the former being eluted faster than the latter (retention volume 10.9 mL vs 9.2 mL, respectively). By using reference proteins the Mr of the two proteins was calculated. In particular, the deglycosylated ovomucoid possessed an apparent Mr of 27 kDa compared to intact ovomucoid with an apparent Mr of 48 kDa (Table 1). We have also calculated the protein Stokes radii as a measure of the hydrodynamic volume of glycosylated and deglycosylated protein forms (Table 1). In fact, the differences in Stokes radii for glycosylated and deglycosylated protein reflect the difference in hydrodynamic volume of each protein form. The Stokes radii of the intact and deglycosylated ovomucoid were equal to 30.8 and 25.6 Ǻ, respectively (Table 1).
Protein |
Mr |
Stokes Radius (Ǻ) |
Intact ovomucoid |
48 |
30.8 |
Deglycosylated ovomucoid |
27 |
25.6 |
Table 1 Mr and Stokes radii for intact and deglycosylated ovomucoid
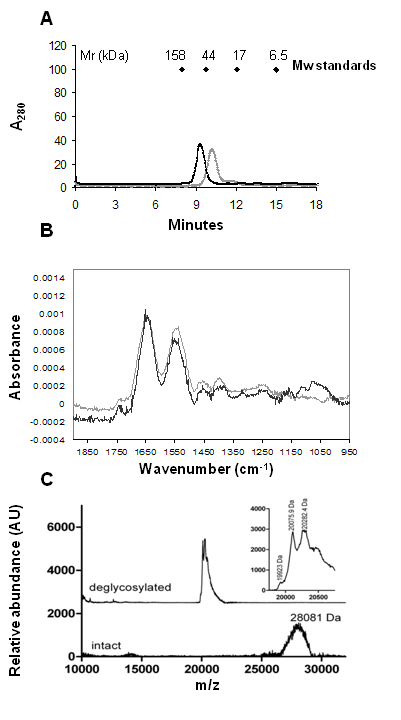
FTIR: The effect of the enzymatic deglycosylation on the intensity of the IR carbohydrate band was also investigated (Figure 5, Panel B). Both the spectra of intact and deglycosylated ovomucoid form appeared quite noisy due to the very low concentration of the protein samples (0.1 mg mL-1). It was not possible to dry down the solution because of the presence of octyl glucoside that interferes with the sugar region, even after exhaustive dialysis of the samples against water. The absorption spectra of the protein before and after enzymatic treatment were normalized at the amide I peak, in the region 1900-900 cm−1 (Figure 5, Panel B). A comparison of the spectra shows differences that are due to the absorption of carbohydrates and clearly evidences a drastic reduction in the band intensity after the enzyme treatment, confirming that the enzymatic treatment removed most of the sugars present on ovomucoid. It is worth pointing out that FTIR spectroscopy could not detect any changes in ovomucoid secondary structure upon deglycosylation, in fact the two protein forms exhibit essentially identical residual amide I and II band shapes, indicating that extensive deglycosylation had little effect on ovomucoid secondary structure.
MALDI-TOF MS analysis of intact and deglycosylated ovomucoid: The deglycosylation reaction was also monitored by MALDI-TOF MS analysis, which can determine the molecular weight of a protein with an accuracy of 0.01%.23 The mass spectra shown in Figure 5, Panel C, demonstrate the mass shift following deglycosylation. In addition, the observed width of the deglycosylated protein peak (Mw ~ 20 kDa) (Figure 5, Panel C) is significantly reduced from that of the native glycoprotein (Mw ~ 28 kDa) (Figure 5; Panel C). This is attributable to the highly heterogeneous glycoforms of the native glycoprotein compared to the PNGase F-treated sample. From the inset shown in Figure 5, Panel C, it is possible to see that the 20 kDa peak is not completely resolved and it is split in two main mass events of 20075 Da and 20282 Da, respectively. The 20282 Da peak most likely represents a modification of the 20075 Da peak. This modification is possibly sinapinic acid adduct formation (+206 Da) caused by the preparation required for MS or could potentially represent acetylhexosaminylation (+203 Da) remaining after deglycosylation of the protein.
LC-MS/MS: In order to look at N-glycosylation sites, bands were cut from a gel (Figure 4); one for the untreated ovomucoid (lane 1) and two bands (at 20 and 22 kDa) in lane 3 corresponding to PNGase F-treated Ovomucoid. These bands were then subjected to in gel digestion using either Trypsin or Chymotrypsin and the resulting peptides analysed by LC-MS/MS to identify the sequences. Using a variable modification for the deamidation of Asn allows detection of sites within a sequence where the PNGase F has cleaved the N-glycans at specific Asn residues. It also allows the detection of unmodified Asn indicating that the Asn has not been glycosylated. The number of modified and unmodified peptides for each N-glycosylation site is summarised in Table 2. Up to eight Asn were found to be potential glycosylation sites for ovomucoid at positions 34, 47, 77, 93, 99, 190, 193 and 199, with peptide sequences identified containing deamidated Asn at these sites in the PNGase F treated bands but not in the untreated ovomucoid band. Some of these sites were not previously identified. In the case of Position 34 twelve and ten different peptide sequences containing the deamidated Asn were identified in the 20 kDa band and 22 kDa bands respectively. The highly variable nature of the glycosylation within the ovomucoid sample analyzed was also demonstrated by the identification of unmodified Asn in all N-glycosylation sites, indicating that at least some of the ovomucoid was not glycosylated at each of these sites. For position 34 six different peptide sequences with unmodified Asn were identified.
P01005* |
PNGase-Treated Ovomucoid 20 kDa Band |
PNGase-Treated Ovomucoid 22 kDa Band |
Untreated Ovomucoid Band |
|||
N-Glycosylation Site |
Peptides Sequences Identified with or without Deamidation Site |
|||||
|
With (Glycosylated) |
Without (not Glycosylated) |
With (Glycosylated) |
Without (not Glycosylated) |
With (Glycosylated) |
Without (not Glycosylated) |
34 |
12 |
6 |
10 |
2 |
0 |
3 |
47 |
1 |
5 |
1 |
3 |
0 |
1 |
77 |
3 |
1 |
0 |
1 |
0 |
1 |
93 |
3 |
11 |
4 |
2 |
0 |
0 |
99 |
4 |
13 |
11 |
1 |
1 |
0 |
190 |
1 |
10 |
0 |
3 |
0 |
4 |
193 |
2 |
6 |
0 |
5 |
2 |
1 |
199 |
5 |
6 |
3 |
2 |
2 |
1 |
Table 2 Number of N-glycosylated and non-glycosylated peptides identified by using LC/MS/MS from in gel digestion of ovomucoid bands
*Gallus gallusovomucoid accession number
An explanation for the two bands obtained for the PNGase F treated ovomucoid can be derived from the mass spectrometry data. Both bands gave consistent data for N-glycosylation sites except in the higher molecular weight band where no peptide sequences with deamidated Asn were identified for Position 77. It is proposed that the reason for higher running band is due to an incomplete deglycosylation at Position 77.
In the last two decades, a wide variety of endoglycosidase and glycoamidase enzymes able to release oligosaccharides from glycoproteins has been discovered and characterized.13-17,24 Using these enzymes it is now possible to conveniently and non-selectively release N-linked oligosaccharides from glycoproteins. Additionally, enzymes that exhibit a surprising degree of specificity with respect to the type of N-linked oligosaccharides released, have now been described and well characterized.17 In this paper an enzymatic strategy, based on deglycosylation by the glycoamidase PNGase F was utilized. This enzyme cleaves the amide bond between Asn and the innermost GlcNac residue on the N-glycan.18 Asn is converted to aspartic acid in this reaction, and glycosylation sites can thus be identified through a mass increase of 1 Da.25
SDS-PAGE analyses have shown that initially the ovomucoid band appeared quite broad, with an apparent Mr of ~ 40 kDa, owing to the heterogeneity of the oligosaccharides as already reported by several authors.8,9,26 However, after deglycosylation using 2.5 U of enzyme, the band became sharp and exhibited an apparent Mr of ~ 20 kDa. This was only observed in the sample that was incubated with PNGase F for 18 h (after only 1 h incubation with deglycosidase, an additional band with a slightly higher Mr of ~ 22 kDa was always observed) following both the heat (100 °C for 1 h) and reducing agent treatments. In fact, the deglycosylation was not complete in either the native form of ovomucoid or the heat-treated and reduced one, even with an excess of PNGase F, as was evident from SDS-PAGE analysis by the formation of faster migrating polypeptides. These polypeptides are likely intermediate glycosylated forms that differ in their degree of deglycosylation.27 Parallel behaviour was observed when samples were analyzed by immunoblot. This result demonstrates that for ovomucoid only the combined effect of the heat and reducing agent treatments makes the protein more accessible to the deglycosidase attack.17,28,29 since only under these conditions is the protein sufficiently unfolded, as we have demonstrated by FTIR and CD analyses. In fact, it is well known that the main problem of protein deglycosylation by glycoamidases is accessibility to the carbohydrate structure by the enzyme. Some glycoproteins are effectively deglycosylated by PNGase F in their native form, but reduction with a sulfhydryl reagent, such as β-mercaptoethanol or DTT, and denaturation with an ionic detergent such as SDS, generally greatly improves oligosaccharide release.16 It is worth pointing out that in our experiments the detergent used for denaturing the protein was octyl glucoside, a non-ionic surfactant, since PNGase F is highly susceptible to denaturation by ionic detergents.17
In order to further characterize the carbohydrate-depleted ovomucoid form, different analytical techniques were applied. One of these techniques was FTIR. The sugar moieties, the protein amide group, and water all produce distinguishable bands in the mid-IR range, and IR spectroscopy has been extensively used to characterize carbohydrates response.2 Our FTIR spectrum showed high intensity in the region 1200-1900 cm-1, corresponding to carbohydrate bands, thus demonstrating the massive presence of sugars on ovomucoid. Comparison of the absorption spectra of the protein before and after enzymatic treatment showed a drastic reduction in the sugar band intensity after PNGase F treatment assessing the removal of most of oligosaccharides. Nevertheless, carbohydrate depletion of ovomucoid did not result in any change of Amide I and Amide II band shapes, suggesting that the sugar moiety does not influence the secondary structure.
In addition, following the complete release of the oligosaccharide from ovomucoid, we could separate the deglycosylated protein and the intact one by passage of the mixture through a column of Superdex 75 gel permeation column under reducing conditions. The deglycosylated protein eluted later than the native glycosylated protein, which is in agreement with its enhanced mobility on SDS-PAGE. In fact, the intact and deglycosylated ovomucoid were eluted as a 48 kDa and 27 kDa protein, respectively. The reduced molecular weight of the deglycosylated ovomucoid compared to intact ovomucoid can thus be attributed to the effect of deglycosylation.30 The gel filtration data were also used to calculate the protein Stokes radii. Deglycosylation resulted in a decrease in Stokes radii from 30.8 to 25.6 Å and a corresponding increase in diffusion coefficients of the intact ovomucoid. These values suggest that there was no change in molecular density as a result of the deglycosylation, which is also in agreement with the spectroscopic data showing no change in secondary structure.
The deglycosylation reaction was also monitored by MALDI-TOF MS analysis. MALDI-TOF MS gives accurate mass, whereas methods, such as SDS-PAGE and gel filtration, only show mobility, which is based on the hydrodynamic radius and not on the mass directly. Our results have shown that upon deglycosylation, the average m/z of ovomucoid is reduced from 28000 Da to 20000 Da. Again we showed that ovomucoid was heavily glycosylated and contained significant heterogeneity in the carbohydrate moiety, as is possible to see from the broad width of the peak corresponding to the intact protein form. Interestingly, the peak corresponding to the intact protein disappeared after the enzymatic treatment, and only a narrower peak could be detected suggesting that the digestion performed by PNGase F was complete. Additional LC-MS/MS performed on both the intact and deglycosylated ovomucoid bands following the trypsin and chymotrypsin in gel digestion revealed notable differences in the fragment ion spectra. A total of 8 potential N-glycosylation sites were uniquely identified Table 2, whereas other authors have identified only 5 sites.8 In fact, we have found three new Asn at position 47, 190 and 193, that are probably glycosylated. Several peptide sequences have been identified as demonstrated by the yield of different identified glycopeptides (Table 2 summarizes the results of this study) confirming the high site-specific glycosylation heterogeneity31,32 existing within the ovomucoid molecule.
In this paper the oligosaccharide fragments were released from the glycoprotein ovomucoid by means of the enzyme PNGase F. The complete denaturation of the protein, by pre-treatments with both heating and reducing agents, was necessary to promote complete access of the cleaving enzyme to the glycosylation sites. In fact, a ladder of partially deglycosylated forms appeared on SDS-PAGE profile if the protein was in its native form or not completely denatured, whereas the extended denaturation led to the formation of a very sharp protein band. Different analytical techniques, such as GF, FTIR and MALDI-TOF, were used to confirm the extent of deglycosylation and asses changes in protein structure resulting from the deglycosylation. Furthermore LC-MS/MS allowed the assignment of new protein N-glycosylation sites, not previously identified. The deglycosylation of ovomucoid is relevant, since ovomucoid protein is the dominant allergen of egg white and the glycans can be involved in its allergenic potential. Furthermore, the strategy shown in this study for a model glycoprotein might be useful for the characterization of other complex food glycoproteins following their deglycosylation.
The authors would like to thank the EU 7th Framework Programme, Marie Curie Intra-European Fellowship (proposal no 235010) awarded to Dr C.V.L. Giosafatto and the UK Biological and Biotechnological Science Research Council through an Institute Strategic Programme Grant to the Institute of Food Research. They also acknowledge the grant from the British Council/Ministero dell’Istruzione, dell' Università e della Ricerca awarded to Dr C.V.L. Giosafatto.
The author declares no conflict of interest.

©2016 Giosafto, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.