MOJ
eISSN: 2381-182X


The Pseudomonas genus is a big problem mainly for the poultry food industry. The shelf life of chicken carcasses stored under refrigeration is limited by the appearance of undesirable odors and sliminess surface generated primarily by Gram negative bacteria. Due to the subsequent emergence of resistant bacteria, is necessary proving new alternatives as guaranty the microbiological quality of foods and human health. Bacteriophages or phages are viruses of bacteria that use resources of bacteria for their replication and killing bacteria “naturally”, showing them as a potential tool for bacteria biocontrol in food industry. In this study, 11 bacteriophages were isolated from the exudate product of defrost of chicken carcasses using as host strains Pseudomonas aeruginosa (ATCC 25619) and Pseudomonas fluorescens (ATCC 13525). This study also aimed at the purification, quantification and morphological and molecular characterization of phages (RFLP). Bacteriophage can be found in all types of environments, it was possible to perform isolation and purification and achieve concentrations of 1011PFU/mL, which demonstrated a broad spectrum of action on different bacteria of the family Enterobacteriaceae. Bacteriophages were classified as belonging to the order of Caudovirales and families Podoviridae and Myoviridae. Bacteriophages showed similar morphology, but some showed different genetic profiles.
Bacteriophagescan be found in the chicken carcasses and the fact that they are regulators of bacterial growth letting their use as antimicrobial agents to prevent spoilage bacteriaas P. fluorescens and pathogens as P. aeruginosaas evidenced in this study. So, bacteriophage with specificity for Pseudomonas can be an interesting instrument for promoting the expansion of the shelf life of chicken products due to their ability to infect this microorganism.
Keywords: lytic bacteriophages, pseudomonas, chicken carcasses, isolation, biocontrol, characterization
Almost every day, many foods are contaminated with pathogen bacteria that demand the use of efficient technology in food industry for consumer safety. Spoilagebacteria cause high losses in food industry and contribute for deteriorating consumer health.1 Therefore, it is necessary to use strict control measures against pathogens and spoilage bacteria.
Pseudomonas spp. are aerobic, non-spore-forming, gram-negative, rod-shaped bacteria.Psychrotrophic pseudomonads are recognized as major spoilage microorganisms based on their extracellular enzymes. The enzymes are generally good indicator of the keeping quality of protein and lipid-rich foods.Although microbial spoilage may merely lead to foodstuffs being rendered unpalatable, it can also result in serious and even fatal illness. These microorganisms are a big problem for the food industry, mainly the poultry industry. The Pseudomonas fluorescens group frequently causes the spoilage of dairy products. It is also important in patients with burns and cystic fibrosis. Pseudomonas aeruginosa causes spoilage of foods and is pathogenic for humans and animals, often as a secondary infection. It is now recognized as a common source of many community acquired and nosocomial infections, and affects primarily immune-compromised people and those suffering from cystic fibrosis. The most common infections involve the cornea, skin, urinary tract, and respiratory tract, although infections may occur in essentially all anatomical locations.2-4 Rapid increase of antibiotic resistance in the bacterial community is an ecological phenomenon, but is also one of the greatest hazards for human and veterinary medicine worldwide. Circulation of antibiotic-resistant bacteria occurs mainly between four ecosystems (humans, animals, soils and aquatic environments) interconnected by water.5 Many aerobic pseudomonads are resistant to several antibacterial agents. This subject is of particular medical importance because members of this group, such as P. aeruginosa, are serious opportunistic human pathogens. The effective antimicrobial agents for the treatment of P. aeruginosa infections include some β-lactams, such as carbenicillin, ticarcillin, third-generation cephalosporins, the synthetic monocyclic β-lactam aztreonam, carbapenems, the aminoglycosides, and the quinolones. Pseudomonas aeruginosa and other fluorescent Pseudomonasspecies are in general resistant to β-lactams.3,5-7
These facts, making necessary novel and most effective approaches for treat infections caused by multidrug-resistant bacteria. In this context, bacteriophages suggest be a potential tool for treated this type of bacteria due to ability for infect and kill bacteria.8 This tool, was a useful practice before the advent of antibiotics, but in nowadays the antimicrobial resistance is increasing and require others strategies to combat. Bacteriophages are considered the most abundant organisms on the planet. It is estimated that the number of phages on the planet are higher than 1030 viral particles.9,10 Phages are considered ubiquitous agents.8,10 Like all virus, phages do not have intrinsic metabolism and depend on metabolic machinery of host bacteria for replication.From one bacterium, almost 100 new viral particles can be released, all capable of infecting new bacteria. So, the infection cycles have potential to remain until all bacteria are dead.4 Even creating a resistance to a particular phage, bacteria is susceptible to other with similar hosts. Therefore, they can be biocontrol agents in food, and a great tool against resistance to antibiotics.11,12 The use of bacteriophages to control pathogens is promising and is becoming a reality. Although the practice of primary bacteriophage therapy has been performed with a view to the treatment of bacterial infections in humans, the concept of removing undesirable bacterial populations using bacteriophages can be extended to animals, plants, foodstuffs and other domains.13
Considering that bacteria are becoming increasingly resistant to antimicrobials, the present work aimed to isolate and characterize lytic bacteriophage for Pseudomonas spp as a potential alternative for decontamination of food products, mainly poultry products.Organisms and growth conditions
The host bacterial strains for phage isolates were P. aeruginosa (ATCC 25619) and P. fluorescens (ATCC 13525). The phages were isolated according to the adapted methodology of Sambrook & Russel14 and Atterbury et al.15 from 11 samples of frozen chicken carcasses, in 15 slaughterhouses in Minas Gerais state in Brazil. Stock cultures were maintained frozen at -20 °C. Pseudomonas grew in optimal conditions (30 °C/24 h) in Brain Heart Infusion medium (BHI-Himedia, Mumbai, India). Pseudomonas had previously been characterized as resistant to amoxicillin, nalidixic acid, meropenem, trimethoprim and aztreonam (SOTO et al, unpublished data).
Table 1 showed other bacterial strains used to evaluate the host range of isolated bacteriophages. All bacteria were subcultures placed on BHI agar plates at optimal temperature for 24 h.
Microorganism |
Bacteriophages |
||||||||||
Pa1 |
Pa2 |
Pa3 |
Pa4 |
Pa5 |
Pa6 |
Pa7 |
Pa8 |
Pa9 |
Pa10 |
Pa11 |
|
Salmonella Enteritidis ATCC13076 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
Klebsiellapneumoniae ATCC10031 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
Salmonella Typhimurium ATCC14028 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
Salmonella Pullorum ATCC9120 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
S. Choleraesuis ATCC10708 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
Salmonella Abony ATCC6071 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
S.arizonae |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
Escherichia coliATCC11229 |
- |
- |
- |
- |
- |
- |
+ |
- |
- |
- |
+ |
S. aureus |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
S. aureus |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
P.aeruginosa ATCC25619 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
P. Fluorescens ATCC13525 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
P. putida |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
Enterobacteraerogenes ATCC13048 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
+ |
Table 1 Evaluation of lytic spectrum of isolated bacteriophages with Pseudomonas aeruginosaand Pseudomonas fluorescens as host, in relation with others bacteria
(+) Positive lysis; (-) Negative lysis
Bacteriophage isolation and purification
The bacteriophages were isolated from chicken frozen carcasses collected from 15 different slaughterhouses located in Minas Gerais state. 10 mL of each sample (chicken defrosted carcasses) were diluted at the proportion of 1:1 in SM buffer (Magnesium sulfate: 50 mmoles•g-1 Tris-HCl (Sigma) [pH 7.5], 0.1 moles•g-1 NaCl (Merck), 8 mmoles•g-1 MgSO4.7H2O (Chemco), 0.01 % gelatin (Difco™) with agitation during 5 minutes.
The suspension was incubated in shaker (Braun-Biotech International) to 150 rpm, 17 °C/24 h so as to allow phage migration to buffer. After incubation time, 8 mL was transferred to the centrifugation tube and 1 mol•g NaCl was added in standby for 15 minutes. Subsequently, it was supplemented with chloroform (Isofar) and the sample was refrigerated for 30 minutes. Finally, the sample was centrifuged to 4.000 g/ 20 min/ 4°C and the supernatant was collected. Each phage isolate was subjected to a minimum of three successive rounds of serial plaque purification and final lysates were stored in SM buffer at 4 °C for further use.
Phage presence determination
To determine if the isolate procedure was correctly performed, it was used the droplet surface technique modified by Hungaro et al.16 The bacterial strains ATCC 25613 and ATCC 13525 were previously activated in TSB (Trypticase Soy Broth – Himedia) 30 °C/24 h. After incubation, each suspension was adjusted to 108 UFC/mL (D.O: 1.0 / 600nm) in a spectrophotometer (Bioespectro - SP22).
In 5 mL of TSA semi-solid agar (0.7% bacteriological agar/100 mL TSB), it was added 0,5 mL of each bacteria, which were added over a petri plate containing TSA (Trypticase Soy Agar-Himedia). After solidification, micro-droplets (0,1 mL) of each phage suspension were deposited over the surface plate. The plates were incubated for 18-24 h/30 °C.
Phage purification and propagation
Phages were purified according to Sambrook & Russel14 methodology, by adding concentrated phage suspension (0.3 mL) to bacterial strains (0.2 mL/DO: 0.5-600nm). Then, they were incubated at 30 °C/30 minutes. Later, the suspension was added to TSA semi-solid agar and the set was supplemented over petri plates containing TSA. The plates were incubated at 30 °C/24h. After incubation time, isolated lysis plates were identified, one was cut and transferred to a tube with bacterial culture (DO: 0.5/600 nm) and incubated again for 24 h/30°C. Next, it was added 1 mol/g NaCl with standby for 15 minutes. Later, it was supplemented with 10% of chloroform (total volume) and the sample was incubated at 4°C/30 minutes. Finally, the resulting suspension was centrifuged (4.000 g/20 min/4°C) and the supernatant was recovered and stored at 4°C.
Phage quantification
Phage stocks were serially diluted in SM buffer to achieve a concentration that would provide individual lysis plaques in a bacterial lawn. Aliquots (100 μL) of diluted phage solution, 100 μL of a bacterial overnight culture (DO: 0.5/600 nm), and 3 mL of semi-solid TSA agar were mixed and poured onto TSA agar plates. The plates were incubated overnight at 30 °C and the number of plaques was counted on the appropriate dilutions, providing between 10 and 100 plaques.
Phage morphologic characterization
The bacteriophage morphology exam was performed as described by Sambrook & Russel.14 Phage suspension (0.1 mL) and 10% of PEG 8000 (Sigma) were transferred to a microtube. The mixture was maintained under refrigeration for 14 h and later centrifuged (11.000 g/20 min/4 °C). The supernatant was discarded and the pellet was resuspended in 1 mL of SM buffer and incubated for 1 h/24°C. Then, chloroform (1 mL) was added and homogenized. The suspension was centrifuged (3.000 g/15 min/4°C) and the supernatant was recovered.
0,8 mL of supernatant was deposited on the transmission electron microscopy (TEM) film provided with carbon-coated Formvar (Koch), stained with 2% uranyl acetate (Sigma-Aldrich, Saint Louis, USA), rinsed with distilled water and allowed to dry for 24 h. After drying, was examined in a Zeiss EM109 transmission electron microscope (performed by Microscope Center, Universidade Federal de Viçosa, Brazil).
Phage lytic spectrum
The host range of each phage was determined against 15 bacterial strains (Table 1). The susceptibility of various bacterial strains was tested using the drop-on-lawn technique.16 Aliquots (10 μL) of serial dilutions of the different phage suspensions were added to bacterial culture. The plates were incubated overnight under optimal condition for each strain, and the lytic activity was checked for the formation of clear areas and phage plaque formation on the bacterial carpet. Each microorganism was incubated at the optimal growth temperature.
DNA extraction and Restriction Fragment Length Polymorphism (RFLP)
Purified phage suspensions were previously treated for 10 minutes with chloroform 50% (Carlo Erba, Italia) and centrifuged (10.000xg/20min). The supernatant was treated with DNase I (Sigma-Aldrich) and RNAse A (1 µg/ml) to disrupt bacterial DNA and RNA. Capsids were disintegrated using proteinase K (Sigma-Aldrich), isopropanol (Vetec) and sodium dodecyl sulfate-SDS (10%) (LCG Biotecnologia). Phages DNA were extracted by the phenol/chloroform method and precipitated by standard ethanol (70% - Vetec) procedure. Phage DNA were extracted using the adapted protocol of Sambrook et al.14
The DNA was digested with HaeIII, HinfI and AluI (Promega) following the manufacturer's recommendations. The DNA extracted was quantified by use of quantity markers λDNA (25, 50 and 100 ng) and running in agarose gel (1.2%) (Invitrogen). For RFLP, it was used DNA molecular marker (1 Kb - Promega) and agarose gel (1.5%). The obtained fragments were visualized after electrophoresis and illumination by UV light (Vilbert Lourmat).
Phage isolation, purification, propagation and quantification
Eleven bacteriophages were isolated and purified successfully from product of chicken carcasses defrost with bacterial culture tested (Figure 1). Initially, these phages were isolatedbased on their ability to lyse P. aeruginosa (ATCC 25619) (phages assigned asPa1 to Pa10) and P. fluorescens (ATCC 13525) (Pa11). The specificity was identified after incubation for 24 h/30°C.
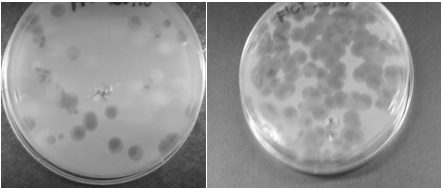
Figure 1 Lysis plaques caused by phages isolated from defrost chicken carcass exudate. That lysis evidenced phage action onto P. aeruginosa and P. fluorescens
The exudate of chicken was an excellent source for phage isolation for these strains, and phage specific for Pseudomonas could be isolated from other sources, such as water, wastewater, feces, soil, and others, provided that a host is present.17,18 During propagation, phages proved capable of replication and achieved concentrations of up to 1011 UFP/mL. Hungaro et al.16 observed that bacteriophages in high concentration (near to 109 PFU/mL) are necessary for optimum efficacy of phage, however, concentrations equal or less than 106 PFU/mL were not able to reduce growth of S. enteritidis in chicken carcasses. So, our bacteriophages for Pseudomonas can be propagated easily and reach high concentrations, this feature, can be considered another advantage for potential use in the food industry.
Phage morphological characterization
Electron transmission microscopy evidenced that the phages isolated in this work can be classified into the Caudovirales order but in different families. All phages showed size approximate of 200 nm. Figure 2 showed the typical morphology of phages, the object of this study.
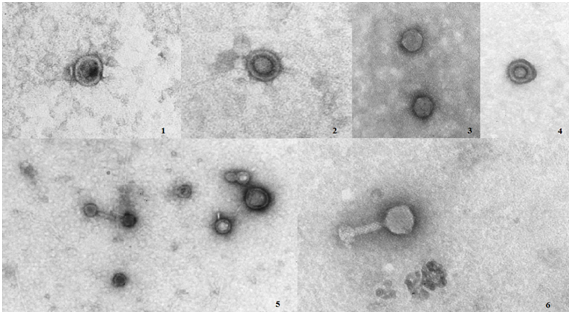
Figure 2 photomicrographs of bacteriophages: 1) Pa3, 2) Pa5, 3) Pa7, 4) Pa8, 5) Pa10 and 6) Pa11, visualized in TEM (8.500X)
Specific phages for P. aeruginosa were included into the Podoviridae family. These phages are characterized by icosahedral heads, dsDNA and non-contractile tail.10
On the other hand, Pa11 phage with specificity for P. fluorescens was included into the Myoviridae family, presented contractile neck10 and dsDNA. Although having similar morphology, the genetic profile of phages is not obligatorily similar, because the environment of isolation is different and faraway. According to Garbe et al.19 499 bacteriophages have been described particularly for Pseudomonas, out of which 139 belong to the Myoviridae family. Kumari et al.18 Sepulveda-Robles et al.20 & Sillankorva et al.21 found similar results for isolated bacteriophages for P. aeruginosa and P. fluorescens.
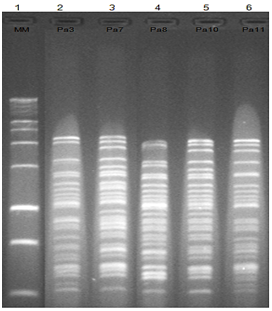
Figure 3 Patterns of digestion with restriction endonucleases HaeIII of DNA isolated phages. Column 1: molecular marker (1 Kb); Column 2-6: DNA phages: Pa3, Pa7, Pa8, Pa10 and Pa11 respectively, digested with Hae III enzyme.
Lytic spectrum
Table 1 exhibited phage activity propagated into P. aeruginosa and P. fluorescens. This analysis is related to phage action on different strains of importance for human and animal health.
PA11 phage showed specificity in many bacteria such as S. Enteritidis, K. pneumoniae, S. Typhimurium, S. Choleraesuis, E. coli, P. aeruginosa, P. Fluorescens, P. putidaandE. aerogenes and almost all these microorganism represented some problem for food industry. Other phage that showed an interesting profile was PA6. Due to action spectrum, these bacteriophages can be interesting candidates for use in biosanitization and phage therapy in food industry. Some phages specific from bacteria belonging to the Enterobacteriacea family were capable of infecting other strains.22,23
The similarity between the compositions of bacterial wall surface of Gram negative bacteria that serve as receptors for bacteriophage ligations can have influence in these results. These bacteriophages were not specific against to Gram positive bacteria tested. Phages with capacity to infect several hosts included different genus of microorganism exhibit potential of use in control and therapy of foodborne pathogens. This characteristic avoids simplification and reduces production costs of phage suspension, also increasing the safety margin, meanwhile the phage used against pathogens could be replicating in non-pathogen bacteria.23
Physical, chemical and biological methods have been studied and applied on food chain to reduce food borne pathogens and spoilage bacteria, especially in the poultry industry. Bacteriophages can be studied and coupled with these methods to improve microbiological safety in food industry. To increase the potential efficiency of this antimicrobial alternative, a mix of phages is an option that could contribute to combatseveral pathogens and antimicrobial resistance.
Genetic profile of bacteriophages
The DNA extracted showed concentration of approximately 50 ng of λDNA marker. It was possible to identify that 5 isolated bacteriophages had different restriction profiles, according to RFLP. The RFLP detected with HaeIII enzyme allowed differentiating phages Pa3, Pa7, Pa8, Pa10 and Pa11. It suggests that isolated phages have different genetic profiles (Figure 3) although of their very similar morphology. The other restriction enzymes tested do not showed fragmentation on bacteriophages DNA, clearly.
Knezevic et al.24 identified and characterized bacteriophages for P. aeruginosa. Based on RFLP profile with EcoRI, EcoRV and BamHI enzymes, they concluded that their phages were genetically different.
Seven phages used in this study presented similar morphology andits restriction digest profiles were indistinguishable from each other.Also, there were not evident differences between of thesephages, their replication dynamics and host range. Sequencing is required to complete identification, although the morphological and genetic characterization of these phages suggests that they belong to identical phage-types.
In the present work, it was observed that phages can be isolated of industrial foods. Bacteriophage PA11 have a big potential for use in food industry as indicate or as control foodborne pathogens and spoilage bacteria, due to their large spectrum of action and their high concentration achieved. The fact of isolated phages of Pseudomonas has a large specificity represent a big advantage and we believe that bacteriophage-based treatment can be applied forpathogen control in poultry carcasses and others industrial products. However, studies are necessary to know the behavior of bacteriophage in food industryfor future applications.
The research group thanked the Núcleo de Microscopiae Microanálisis of University of Viçosa-Minas Gerais, Brazil for their help in the determination of phage morphology.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.