Journal of
eISSN: 2373-4310


Review Article Volume 2 Issue 1
Department of Health, Baylor University, USA
Correspondence: Darryn Willoughby, Department of Health, Human Performance, and Recreation, Baylor University, Waco, TX 76798, USA, Tel (254) 710-3504
Received: December 12, 2014 | Published: February 9, 2015
Citation: Hwang P, Willoughby DS. Intracellular sensing and transport of amino acids and the downstream effects on mtorc1 activation: implications for muscle protein synthesis. J Nutr Health Food Eng. 2015;2(1):25-33. DOI: 10.15406/jnhfe.2015.02.00044
It is widely established that the rate of muscle protein synthesis is elevated with amino acid availability. However, when there is a lack in amino acid availability homeostatic processes are implemented in order to prioritize the energy demands necessitating amino acid scarcity. To this respect, it is understood that amino acid availability is a rate-limiting factor underlying the mediation of muscle protein synthesis (MPS). The mammalian target of rapamycin complex 1 (mTORC1) is a master regulatory complex that plays a significant role in meticulously coordinating the myriad of signaling cascades in order to up-regulate MPS activity through increased protein translation initiation. The presence of adequate amino acids such as leucine is evidenced to be necessary for the activation of mTORC1. Moreover, amino acids have been known as subsequent mediators that trigger mTORC1 activation independent of the traditional upstream signaling cascades associated with growth factors. It has recently been observed that the activation of mTORC1 is implemented at the surface of the lysosome membrane. As a result, considerable advances have occurred which demonstrate various protein components which relay on amino acids to relay signals to mTORC1. This review will focalize upon describing the present understanding of the multiple signaling components that mediate signals in conditions of adequate amino acid levels to mTORC1. Furthermore, this review will also present recent advancements behind amino acid sensing mechanisms through the discussion of upstream components of mTORC1, amino acid transporters, as well as proposed underlying mechanisms of intracellular signaling relative to amino acid sensing.
Keywords: muscle protein synthesis, amino acids, leucine, rag gtpase, mtorc1, lysosome
MPS, muscle protein synthesis; mTORC1, mammalian target of rapamycin complex 1; mTOR, mammalian target of rapamycin; PI3K, phosphatidylinositol-3 kinase; PIP2, phosphatidylinositol-4,5-biphosphate; PIP3, phosphatidylinositol-3,4,5-triphosphate; PDK, phosphoinositide-dependent kinase; 5’-TOP, 5’ terminal oligopyrimidine; EIFS, eukaryotic initiation factors; GAP, gtpase activating protein; BCAAS, branched chain amino acids; HBXIP, hepatitis B virus x interacting protein; GEF, guanine nucleotide exchange factor; PA, phosphatidic acid; PLD1, phospholipase D; GDH, glutamine dehydrogenase; IPMK, inositol polyphosphate multi-kinase; ULK1, unc-51-like kinase 1
The mammalian target of rapamycin (mTOR) signaling pathway has attracted considerable attention due to its involvement in regulating muscle protein synthesis (MPS).1,2 Additionally, this protein kinase is also involved in regulating various intramuscular and developmental processes such as gene transcription, mRNA translation, cell proliferation, metabolism, and repression of autophagy.3 There is substantial evidence demonstrating that mTOR is similar to a “master switch” mediating the balance between intramuscular anabolic and catabolic processes due to its complex ability to sense mitogen, energy, and nutrient levels.4,5 Several studies have demonstrated that amino acids, such as L-leucine, or heightened activity of mitogen activity (growth factors such as IGF-1 or insulin) can activate the signaling cascade of mTOR for subsequent increases in protein translation initiation. Furthermore, concomitant to this augmented rate of protein translation initiation, skeletal muscle protein turnover can also increase the positive net protein balance over time.6 However, it is generally understood that these mitogens seem to act upon different substrates upstream or downstream from mTOR. With respect to the mTOR signaling pathway, growth factors such as insulin or IGF-1 are known to bind their respective receptors, which would trigger the activation of various kinases, phosphatidylinositol-3 kinase (PI3K) in particular.4,7 Activated PI3K catalyzes the conversion of membrane phospholipid phosphatidylinositol-4,5-biphosphate (PIP2) into phosphatidylinositol-3,4,5-triphosphate (PIP3), which would open a lipid-binding membrane site for effector protein kinase B (Akt).7,8 The increased levels of PIP3, and activity of phosphoinositide-dependent kinase 1 (PDK1) and PDK2, lead to the activation of Akt.5,9 Therefore, this growth factor signaling cascade culminates in a positive regulatory function of mTOR due to activated levels of PI3K and Akt.
Activation of mTORC1 results in phosphorylation of the downstream effectors, eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) and the phosphorylated ribosomal protein S6 (p70S6K). As such, 4E-BP1 is known as an inhibitor of the mRNA cap-binding protein eIF4E.5,8,10 Moreover, the stimulation of 4E-BP1 results in the release of eukaryotic initiation factor 4E (eIF4E), which subsequently enables the recruitment to the second initiation factor, eIF4G.11‒15 This formulates into the multi-protein complex known as the eIF4F translation initiation complex, which functions to assist in the binding of mRNA towards the 40S ribosomal subunit. The enhanced assembly of this eIF4F complex is a significant step for the promotion of cap-dependent protein initiation to ensue.5,10,11,16
The mTOR pathway can also activate p70S6K, which is known as a crucial mediator for the MPS signaling cascade.11 The activation of this signaling intermediate can then phosphorylate its effector ribosomal protein S6 (rpS6), which mediates the enhancement of the protein translation from template mRNA.5,8,10,17 The activation of p70S6K and its effector rpS6 can also enhance translation of mRNAs containing the 5’ terminal oligopyrimidine tract (5’-TOP) (encoding elongation factors as well as ribosomal proteins), consequently increasing protein translation capacity.5,8,10,12,18 In addition, the activation of p70S6K assists in bringing rpS6 in closer proximity towards the eukaryotic initiation factors (eIFs) and mRNA.11 The active p70S6K may also target eIF4B at Ser422, which favors its binding onto eIF4A, leading to increases in the helicase activity of eIF4A, and also providing an additional stimulus for the initiation of protein synthesis to occur.13,14
mTOR is known to function as two distinct multi-protein signaling complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), which differ in their subunit composition, respective sensitivity to rapamycin, upstream integrative patterns, regulation of substrates, and the control of biological processes.5,19‒21 The main function of mTORC1 is to mediate processes of cellular growth, proliferation and survival while promoting ribosomal biogenesis, lipid and nucleotide synthesis, and inhibiting autophagy (Figure 1).5,19 The mTORC1 complex consists of mTOR, regulatory association protein of mTOR (Raptor), mammalian lethal with SEC13 protein 8 (mLST8), Akt/PKB substrate 40 kDa (PRAS40), and DEP domain containing mTOR-interacting protein (Deptor).19 Raptor is known as the essential scaffolding protein component of mTORC1, which functions to recruit and phosphorylate the pertinent downstream substrates, p70S6K and 4E-BP1, in order to mediate changes in the rate of protein translation initiation.5,19
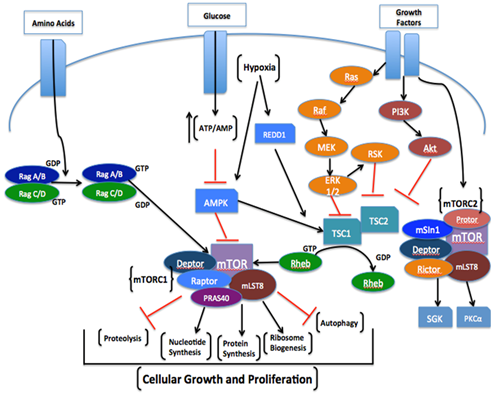
Figure 1 Comprehensive illustration of the mTOR-signaling pathway in which the key signaling pathways involved in mTORC1 and mTORC2 regulation are depicted. As can be seen, multiple inputs from growth factors (i.e., insulin and IGF-1), amino acids (i.e., leucine), intracellular energy status resulting from glucose uptake, and cell stress from hypoxia are integrated into mTORC1. The up-regulation of mTORC1 plays a key role in promoting cell growth and proliferation by stimulating various anabolic processes such as MPS, nucleotide synthesis, and ribosome biogenesis while concomitantly inhibiting processes such as muscle proteolysis and autophagy. Regarding mTORC2, it is regulated by growth factors but through a poorly understood mechanism. However, unlike mTORC1 it does not respond to other up-stream nutrient-derived signals or cell stress [19].
The mTORC1 complex consists of mTOR, regulatory association protein of mTOR (Raptor), mammalian lethal with SEC13 protein 8 (mLST8), Akt/PKB substrate 40 kDa (PRAS40), and DEP domain containing mTOR-interacting protein (Deptor).19 Raptor is known as the essential scaffolding protein component of mTORC1, which functions to recruit and phosphorylate the pertinent downstream substrates, p70S6K and 4E-BP1, in order to mediate changes in the rate of protein translation initiation.5,19 mLST8 is identified as a positive regulator of mTOR kinase activity and is suggested to be necessary for amino acid and rapamycin interaction between raptor and mTOR. However, further research is needed to clarify the potential role of mLST8 within the mTORC1 complex. PRAS40 is clearly identified as a negative regulator of mTORC1 activity.5 To this respect, PRAS40 is a substrate for Akt and the 14-3-3 binding protein.5,21 The activation of the PRAS40 subunit can inhibit mTORC1 activity by functioning to prevent Raptor from recruiting the downstream effectors of mTORC1.22,23 The Deptor subunit is suggested to also function as a negative regulator of mTORC1 activity. By comparing both mTOR complexes, it is suggested that mTORC1 is highly sensitive to rapamycin in comparison to mTORC2, and is more directly involved in up-regulation MPS. Presently, it is understood that mTORC1 activity is mediated by a variety of upstream signals including growth factors and nutrients such as amino acids and glucose.19 Growth factor associated mTORC1 activation is known to lead into the direct activation of the PI3K/Akt and Ras-MAPK signaling cascades.19,24 Furthermore, these particular pathways are regulated by the phosphorylation and inhibition of the tumor suppressor tuberous sclerosis complex ½(TSC 1/2).21,25 This complex is known as an active dimer that negatively mediates mTORC1 signaling through its respective GTPase activating protein (GAP) activity upon small GTPase Rheb (Ras homolog enriched in brain).5,19,21 When the TSC 1/2 complex is phosphorylated by Akt, the function of the TSC2-GAP activity is repressed, thereby ensuring that Rheb maintains its active GTP-bound form. Consequently, growth factors such as insulin or IGF-1, activating their respective trans-membrane receptors, can up-regulate the PI3K/Akt signaling cascade and phosphorylate the TSC 1/2 complex, which will up-regulate mTORC1 activity by elevating the Rheb-GTP (active) to Rheb-GDP ratio.19 Moreover, when Rheb is in its active GTP-bound state, it translocates to the lysosome where mTORC1 is mobilized for subsequent activation.5
It is widely understood that the proper balance between anabolic and catabolic processes is intricately coordinated by both the meticulous implementation of energy expenditure and input. Furthermore, adequate amino acid availability will favor a net protein balance, resulting in an augmentation in MPS and concomitant suppression of proteolysis. These anabolic adaptations can be further augmented by the integration of amino acid supplementation. To this respect, studies have shown that the ingestion of branched chain amino acids (BCAAs), especially leucine, can lead to a prominent increase in the phosphorylation of biomarkers such as p70S6K, 4E-BP1 and rpS6.26‒29 Additional studies have also observed that a combination of resistance exercise with dietary amino acids, primarily leucine, can elevate MPS during the post-exercise recovery period.30‒33
Leucine is known as a potent activator of MPS by playing a major role in activating the mTOR signaling pathway, independent of the traditional growth factor receptor-mediated signaling cascade. In addition, there is evidence demonstrating that leucine can repress ATP-dependent proteolytic pathway-mediated degradation and augment mTORC1 signaling.34 However, there have been a few conflicting studies observing no significant additive differential effect in MPS within subjects provided adequate whey protein versus an enriched whey and leucine supplement prior to resistance exercise.34,35 Nevertheless, there are still a number of studies presenting significant evidence of positive effects on MPS with leucine supplementation.34,36 Moreover, the elevation in leucine availability may augment mRNA translation through the alteration in activity of signaling intermediates within the mTOR signaling cascade.30,37 Based on this reasoning, the significance of amino acids (leucine in particular) and MPS may be further substantiated by elucidating the mechanisms of action behind amino acid availability and mTOR pathway activity.
Amino acid availability is crucial for proper cell growth. Furthermore, through the implementation of energy-consuming anabolic pathways with the integration of adequate nutrient sufficiency, processes of MPS can ensue. Since the mTOR pathway mediates these crucial processes associated with metabolism and cellular growth, the identification of the myriad of input signals are of importance. It is generally understood that amino acids may activate mTOR independently of the traditional growth factor activated PI3K/Akt/mTOR signaling pathway.38 According to the literature, the mechanisms involved with amino acid regulation of MPS may not require the activation of Akt.39 Furthermore, it is suggested that the presence of amino acids assists in regulating the intracellular localization of mTORC1 to the lysosome membrane where mediated association with Rheb may occur for subsequent activation (Figure 2).40 Although it is well understood that amino acid availability is up-regulates mTORC1 activation, there have been a lack of clarity behind understanding how amino acids actually regulate mTORC1.

Figure 2 The alleged amino acid sensing mechanistic pathway for mTORC1 activation.(A) During amino acid insufficiency, Ragulator is resides in an inhibitory state with the v-ATPase, and GATOR1 [GAP activity toward Rags (Ras-related GTPases)] induces activity of GTPases-activating protein (GAP) towards Rag A, thereby maintaining GTAases in its inactive GDP-bound state and unable to recruit mTORC1. Insulin signaling has an inhibitory effect on the tuberous sclerosis complex (TSC) translocation to the lysosomal surface where it functions as a GAP for Rheb (Ras homolog enriched in brain), thereby causing G-protein inactivation. (B)During amino acid sufficiency, GATOR1 can be inhibited by GATOR2. Furthermore, Ragulator and v-ATPase undergo a conformational change, thereby unleashing the activity of the guanine nucleotide exchange factor (GEF) towards Rag A, while the folliculin complex promotes Rag C GTP hydrolysis. This results in an active heterodimer, which consists of GTP-bound RagA and GDP-loaded Rag C and is able to recruit mTORC1 to the lysosomal surface where it interacts with, and is activated by, Rheb [40].
In accordance with the amino acid-dependent mechanism action of mTORC1, researchers have presented compelling evidence on the Rag family of GTPases and utilized strategic RNAi and biochemical methodologies to discover the significance of the Rag GTPases as regulators of amino acid signaling-induced mTORC1 activation.19,41,42 Currently, it is understood that there are four Rag GTPase proteins within mammals (novel family of small GTPases: A-D) that are constitutive heterodimers through which the depletion of any Rag heterodimer may inhibit mTORC1 signaling.43 Rag A and B are similar in amino acid sequence and functionally redundant, as are RagC and RagD.19 Moreover, extracellular amino acid transport across the plasma membrane is necessary to activate mTORC1.40,44 Furthermore, in the absence of extracellular amino acids, the utilization of a cell-free reconstitution assay suggested that the location of the amino acid signaling might occur within the lysosomal lumen. Moreover, the depletion of the intracellular lysosomal amino acid levels would theoretically repress the amino acid-dependent recruitment of mTORC1 to the lysosome for activation.44,45 The Rag GTPases, which reside downstream of the amino acids, would then function to communicate their availability to mTORC1.44 The Rag GTPases localize to the lysosome membrane surface where they function to recruit Raptor in an amino-acid dependent manner, which potentially bridges with mTORC1 activation.44,46 The nucleotide-bound state of the Rag GTPase heterodimers is essential for amino-acid dependent mTORC1 recruitment to occur. Rag GTPase A/B binds to GDP during conditions of amino acid scarcity, which is consequently changed to a GTP-bound state following the repletion of amino acid levels.40,44,46‒48 This is also confirmed in other studies reporting that the expression of a dominant-inactive mutant form of Rag A and B fixated in a guanine nucleotide-free form nearly repressed mTORC1 activity even during conditions of amino acid sufficiency.19,42,48 These studies also found that the dominant-active mutant of RagA and B in active GTP-bound form ensured the maintenance of mTORC1 activity during conditions of amino acid starvation. Interestingly, the expression of mutants for RagC and D did not significantly affect mTORC1 activity. Based on these observations, it can be inferred that amino acids mediate the nucleotide-bound state of RagA and B while this particular dimer pair carries precedence as the major determinant in mTORC1 activation at the lysosome.19 It is important to note that RagC and D are still necessary for mTORC1 activity by forming the stabilized heterodimer conformation with RagA and B for enhanced expression.
Another recent study has also observed that an intracellular signaling adaptor molecule known as p62 functions as a Raptor interacting protein necessary for amino acid-dependent activation of mTORC1.34,49 Moreover, it was suggested that p62 is a crucial mediator of Rag signaling to mTORC1 for its respective localization to the lysosomal membrane to be activated. However, a limitation in this study included the lack of findings investigating whether this signaling adaptor molecule has an interaction with the Ragulator complex (which is important for Rag-GTPase-mTORC1 localization to the lysosomal membrane). Nevertheless, further research is needed to confirm whether p62 plays a significant role in both amino acid sensing and Rag GTPase signaling to mTORC1. According to the literature, it has been observed that during conditions of sufficient amino acids levels and expression of the active RagA/B conformation, the active Rag complex was able to bind to the raptor subunit of mTORC1 and assist in recruiting mTORC1 to the lysosome to interact with activated Rheb-GTP.19,42,43 Based on these findings, a model has been proposed delineating the role of amino acids mediating the GTP state of RagA and B, which would enable the convergence of pathways including the amino acid-Rag and the growth factor-Rheb interaction with mTORC1 localization to the lysosome for efficacious activation.19,42,50 However, further research is necessary to elucidate and confirm the spatial regulation of mTORC1 between the Rag GTPases as well as the localization steps for endogenous Rheb at the lysosome organelle, respectively.
The Rag GTPases do not contain lipid modifications, which are utilized to target intracellular proteins. Therefore, the Rag GTPases may not be able to directly recruit mTORC1 at the lysosomal membrane. In light of this, researchers have discovered the presence of a complex of proteins that function to interact and assist in targeting the Rags onto the lysosomal surface.19,50 This complex is known as the “Ragulator” complex, and functions to anchor the Rag-mTORC1 complex onto the lysosome surface.19 The Ragulator complex was originally thought to exist as a trimeric protein complex comprised of p14, p18, and MP1 (also known as lysosomal adaptor and MAPK and mTOR activator of LAMTOR 1-3).50 However, recent studies have discovered that this complex is in actuality a pentameric complex that also includes C7orf59 (LAMTOR 4) and hepatitis B virus X interacting protein (HBXIP or LAMTOR 5).40 According to the literature, the lipid modification of the Ragulator subunit p18, through reactions of N-terminal myristoylation and palmitoylation, is suggested to anchor this Ragulator complex at the lysosomal membrane surface.19,41,50 It should be noted that repression or genetic deletion of any subunit in this pentameric complex seems to inhibit amino acid-dependent lysosomal localization of Rag, as well as the subsequent movement of mTORC1 onto the lysosome.19 In the presence of amino acids, proper functioning of this whole pentameric complex seems to be necessary for the anchoring of the Rag GTPase complex to the lysosomal surface alongside the consequential activation of mTORC1. In addition, in conditions of amino acid sufficiency, the Ragulator complex is also known to function as a guanine nucleotide exchange factor (GEF) towards the Rag A and B dimers.19,41 This indicates that the exchange of GDP with GTP can be initiated with the Rag A and B GTPases, which may subsequently activate these dimers within this amino acid signaling cascade. Furthermore, it is suggested that the Ragulator complex seems to be bound to the Rag GTPases in their inactive state, which is strengthened by the nucleotide state of the Rag A and B dimers.40,51 In other words, the strength of this Rag-Ragulator interaction seems to be highly dependent upon amino acid levels; Rag binding affinity towards the Ragulator complex is reduced in conditions of amino acid deficiency. Therefore, in conditions of amino acid sufficiency, the Rags are activated through the GEF auxiliary function of the Ragulator complex, which decreases the binding affinity and subsequent ability for the Rags to provide a domain at which mTORC1 can be recognized and localized onto the lysosome.19
Presently, a considerable amount of research has been carried out in order to elucidate the mechanisms underlying the process of amino acid signal transmission and subsequent activation of mTORC1 through the interaction with the Rag GTPases. However, the actual location at which the amino acids are sensed upstream of the Rag GTPases, as well as how exactly the amino acid sensing is initiated, lack clarity. As stated previously, in the presence of amino acids, the Rag GTPase heterodimers are activated alongside the GEF activity of the Ragulator complex, which culminates into the localization and subsequent activation of mTORC1 at the lysosome surface membrane. Based on these observations, researchers have proposed an “inside-out” model such that the lysosomal lumen is the original location in which amino acid sensing may occur.19,50 Moreover, the accretion of amino acids within the lysosomal lumen is suggested to provoke the initiation of sensing signals, which would be mediated through the presence of the vacuolar H+ adenosine triphosphatase (v-ATPase); this would then lead to the subsequent activation of mTORC1 through the active Rag-Ragulator pathway.19,46 This v-ATPase is comprised of two major multi-protein complexes, the V1 and V0 domains, which function in the acidification of the lysosomal lumen.40,41 In other words, this component operates to maintain an optimal low pH in order for the lysosome to function properly. Furthermore, the V1 domain functions by hydrolyzing ATP, which then rotates the V0 membrane domain in order to propel protons (H+) across the plasma membrane into the lysosomal lumen and lower its respective pH levels.19,41,52
The depletion of the v-ATPase subunits seemed to inhibit mTORC1 activation as well as its amino acid-stimulated lysosomal localization, thereby highlighting its significant role in the amino acid signaling pathway for mTORC1 activation. According to the literature, the v-ATPase is suggested to engage in amino-acid dependent interactions with the pentameric Ragulator complex.19,40,46 Higher levels of amino acids seem to lower the binding affinity between only the V1 domain of the v-ATPase and the Ragulator complex.19 Furthermore, the chemical inhibition of this v-ATPase complex seems to repress amino acids from mediating the interaction with the Ragulator complex, which may further confirm the interactive role between this v-ATPase and the Ragulator complex.19 Therefore, the proper functioning of the v-ATPase in the presence of amino acids may bridge the relationship between the GEF activities of the Ragulator complex and the Rags, and their mechanistic role in the amino acid-sensing pathway. However, an important question of relevance may include whether there is a conformational change between the v-ATPase and the Ragulator complex in the presence of amino acid levels, which solidifies the interactive role between these complexes. If there were substantial evidence to support this interaction, an additional question would include whether the v-ATPase complex is a true amino acid sensor within this amino acid-sensing pathway. Interestingly this lysosomal “inside-out” model was also further confirmed by a study which observed that amino acids appear to be sensed within the lysosomal lumen through the respective initiative signal relayed to the v-ATPase and, ultimately, mTORC1 activation.46
Another important issue for discussion is whether amino acid sensing is linked to the amino acid transporter activity which is necessary for amino acids accumulation within the lysosome. If so, this may confer credence to the “inside-out” model previously described. In line with this particular issue, studies have identified various amino acid transporters at the lysosome, which may assist in mediating mTORC1 activity.41 Amino acid transporters may confer an integral role in amino acid delivery to specific intracellular sensors in concordance to the amino acid sensing pathway for mTORC1 activation. With respect to the availability of specific amino acids, leucine functions as a signaling molecule that can directly activate mTORC1 in an Akt-independent manner. Leucine can be transported into the cell via the membrane-bound receptor SLC7A5 (ASC) coupled with SLC3A2 (LAT1/CD98), which also requires the active transportation of glutamine out of the cell.34,53 This presents compelling evidence regarding the significance of glutamine for leucine-induced activation of mTORC1. Accordingly, this particular role of glutamine may, at least partially, explain why it is the most abundant amino acid within the body; this is especially true considering the content of glutamine within skeletal muscle.34
Intracellular leucine homeostasis and the rate of uptake can also be coupled with transporters such as the system L and A transporter (LAT1 and SNAT2, respectively), which may be associated with mTORC1 activation.34 These transporters primarily function to transport leucine in exchange for intracellular glutamine. The LAT1 transporter couples with the glycoprotein CD98 alongside the active sodium-linked SNAT2 transporter. SNAT2 functions primarily to regulate glutamine levels within the muscle. LAT1 functions to expel glutamine in exchange for the uptake of leucine whereas SNAT2 continues to maintain high intracellular glutamine concentrations in order to continue to prolong LAT1 function to up-regulate leucine uptake into the cell.34,54 Various studies have been implemented to characterize the expression of amino acid transporters within human skeletal muscle in conditions of amino acid sufficiency. Moreover, the utilization of these amino acid transporter systems seem to be important in order to facilitate and strengthen the sensing of amino acid levels through the integration of various inputs associated with intracellular and extracellular amino acid availability.55 Another particular transporter known as the protein-assisted SLC36 AA transporter (PATs; PAT1 in mammals) seems to be necessary for mTORC1 activation and cellular proliferation.41,56,57 Furthermore, the PAT1 transporter is suggested to export amino acids from the lysosomal lumen into the cytosol.41,57 In accordance to this study, researchers have proposed a model that suggests the formulation of a complex known as the nutrisome, which is comprised of PAT1, Rag GTPase, Ragulator complex, and v-ATPase, which may work together to activate mTORC1.41 However, the PAT1 amino acid transporter seems to be very specific for alanine, glycine, and proline, which necessitates additional elucidation with respect to amino acid signaling for specific amino acids.41,57 Nevertheless, this model suggests that v-ATPase pumps protons into the lysosome, thereby activating the localized PAT1 to pump amino acids out of the lysosome, which may activate mTORC1.41,58 However, a recent study found that continual expression of the PAT1 would reduce amino-acid dependent mTORC1 activation due to the scarcity of amino acids within the lysosome. In light of this, increasing the expression of active GTP-bound Rag A and B, which evidently recovered mTORC1 activity levels regardless of amino acid conditions, can attenuate the repression. Furthermore, lysosomal membrane permeabilization enables amino acids within the lumen to leave, which was shown to inhibit Rag GTPase-mTORC1 interaction.44,46
Currently, there is still a lack of understanding of the direct mechanism(s) in which amino acids (such as leucine) can activate the Rag/Ragulator complex for the translocation and consequent activation of mTORC1. In light of this, a study proposed that in the presence of leucine, leucyl-tRNA (LRS) relocates onto the lysosomal membrane and directly interacts with this bound amino acid by the Rag GTPases (in particular RagD) and may allow for the formation of the Rag GTPase complex for translocation and activation of mTORC1.45 It was also observed that the inhibition of LRS prevented the activation of mTORC1 even with leucine availability, which highlights the possibility of leucine/LRS activation of GAP activity upon GTPase RagD and lysosomal recruitment for mTORC1 activation.45,55,59 LRS is known as a cytoplasmic enzyme that functions to catalyze the ATP-dependent ligation of leucine to its respective tRNA, and is necessary for MPS to be triggered.44,45,60 Nevertheless, future work is needed in order to verify if the LRS mechanism is a major component in the amino acid sensing pathway for mTORC1 activation. Furthermore, studies have also found that LRS sensing may also occur within the cytoplasm and plasma membrane, which infers that multiple mechanisms for amino acid sensing pathways for mTORC1 may be present. Therefore, future research should be implemented with respect to the validity for the mechanism of action behind LRS and leucine availability.44,45,60
Recently, the leucine binding proteins known as UBR1 and UBR2 were identified, which function as negative regulators of mTORC1 activity.34,61 Furthermore, these proteins are classified as E3 ubiquitin ligases, which function to target specific proteins for the process of ubiquitination and degradation by the 26S proteasome.61 In relation to these binding proteins, a study observed that the over expression of both UBR1 and UBR2 were sufficient to minimize mTORC1 signal activity. In stark contrast, the presence of a high concentration of leucine attenuated the suppression of mTORC1 activity. Therefore, it is conceivable that leucine binds the substrate recognition site on UBR2, which may prevent normal degradation processes to occur.62 In view of this finding, it can be further implied that this attenuation of mTORC1 activity may promote further activation for MPS.
Presently, several potential signaling components have been implicated in playing a role in mediating amino acid-associated activation of mTORC1.34 Recently, Vps34, which is in the class III PI3K, was suggested to be associated with the signal transduction pathway due to the presence of amino acids for mTORC1 activation.5,34 Studies have shown that increasing the expression of Vps34 elevated the rate of mTORC1 signal transduction activity.34,63 Also, a few studies have presented evidence supporting the positive regulatory role of activated hVps34 in a calcium-dependent manner with the condition of adequate amino acid availability.4,64 Interestingly, it was also observed that the binding of calcium toVps34 increased the lipid kinase activity and subsequent mTORC1 signaling activation.34,64 Based on this finding, it was suggested that Vps34 might displace amino acids to serve as a calcium sensor. Future research may be necessary to explore whether Vps34 may be activated specifically in the presence of calcium or amino acids. With respect to the signaling cascade of mTORC1, Vps34 is suggested to relay amino acid signals through the PLD1 axis in order to initiate steps for lysosomal translocation, activation and consequently mTORC1 activation.65‒67 Therefore, it can be implicated that the hVps34-PLD1 axis may serve as another avenue through which amino acids can relay signals to activate mTORC1. Currently, it has been found that Rheb may bind to PLD1 in a GTP-dependent manner and promote mTORC1 activation through the mitogen activated TSC1/2-Rheb pathway.66,68 Moreover, recent findings suggest that phosphatidic acid (PA), a product of phospholipase D1 (PLD1), might also be a potential player in the mediation of the metabolic signaling cascade relative to the hVps34-PLD-mTORC1 pathway.67
It was also reported that another component known as RalA, which is a subunit of the Ras family GTPases, is necessary in order for amino acid-dependent mTORC1 to be implemented.19,65,68,69 RalA is known to play a role in modulating mRNA transcription, membrane transport, cell migration, and amino acid regulation of mTORC1.34,70 Maehama et al.,69 observed that the removal of RalA induced the inhibition of both glucose and amino acid-dependent phosphorylation of the downstream effector, S6K1.34 Furthermore, it was suggested that RalA may be downstream of Rheb and is activated in an amino-acid dependent manner. To this respect, it was implied that RalA may act in an amino-acid dependent pathway downstream of Rheb to activate mTORC1, however, mechanistic details of this pathway necessitates further elucidation.
An important kinase known as mitogen-activated protein kinase 3(MAP4K3), has been confirmed as a putative amino acid sensor and regulator of mTORC1 activity.4,34,71,72 Schriever et al.,72 observed that MAP4K3 was prone to regulatory action in conditions of amino acid deficiency. Moreover, following amino acid starvation, leucine-induced S6K1 phosphorylation was reduced within MAP4K3 knockdown cells, which highlights the implication that this protein may have a role in regulating leucine-mediated mTORC1 activation within human fibroblast cells. Therefore, MAP4K3 may serve as a mediator alongside the Rag-GTPases for the mechanism of amino acid signaling for mTORC1 activation.72 Findlay et al.,71 also observed that MAP4K3 over expression increases mTORC1 signaling activity as evidenced by the up-regulation of downstream effectors S6K1 and 4E-BP1. The authors suggested that this kinase functions through amino acid regulation and is localized upstream of mTORC1.34,71 Recent evidence also shows that the MAP4K3 status of phosphorylated activity may be mediated by the competitive inhibition between a specific PP2A (protein phosphatase 2A) and amino acids.34 Nevertheless, these findings still necessitate considerable investigation in order to confirm how amino acids truly mediate the activity of MAP4K3 as well as how this protein kinase functions within the amino acid signaling cascade for mTORC1 activation.
Glutamine is abundant in the blood and, in skeletal muscle, is metabolized in the mitochondria by glutaminolysis.73 Presently, it is understood that glutamine serves an important role in cell growth, which would implicate associations with mTORC1 activity. To this respect, researchers have identified the importance of glutamine transporters, which are necessary for the mediation of leucine transport into the cell in order to trigger mTORC1 activation. As such, it could be suggested that glutamine may indirectly promote mTORC1 activity as a free amino acid due to its relevance for the uptake of essential amino acids such as leucine. It is also noted that the process of glutaminolysis could allow for the leucine-dependent activation of the RagB heterodimer. This process of glutaminolysis is defined as the conversion of glutamine into alpha-ketoglutarate (alphaKG), which is catalyzed by the enzyme glutamine dehydrogenase (GDH). Furthermore, leucine is suggested to allosterically assist in the activation of GDH to convert glutamine into alphaKG for the Rag heterodimer loading phase to be activated.74 Interestingly, chemical inhibitors or depletion of substrates to produce alphaKG has been shown to inhibit glutaminolysis, which consequently decreased processes of activating expressed RagB and translocating mTORC1 to the lysosome. These events further highlight the potentiated role that glutaminolysis and alphaKG production may serve to assist in leucine-mediated Rag-mTORC1 signaling.19 However, it is important to note that long-term glutamine deprivation resulted in a miniscule inhibition on the processes of mTORC1 activation.19,74 Nevertheless, the novel mechanisms associated with glutamine require greater understanding in order to examine how alphaKG can activate the Rag GTPases for its functional role in the translocation of mTORC1 for consequential activation.
Inositol polyphosphate multi-kinase (IPMK) is known as a signaling molecule with inositol phosphate and lipid kinase activity, which was suggested to play a role in amino acid signaling to mTORC1.19,34,75 A study by Kim et al.,75 investigated the role of IPMK on mediating amino acid signaling to mTORC1. The authors found that in conditions of amino acid availability, IPMK acted as a cofactor for mTORC1 through its function of stabilizing the subunit interaction of mTOR and Raptor. Furthermore, IPMK was suggested to facilitate this interaction in the presence of amino acids, which allowed for the Rag GTPase subunits to bind and continue the process of mTORC1 localization to the lysosome.75 Similar to the importance of the subunit mLST8, IPMK was implicated to mediate amino acid-sensitive interactions between the mTOR-Raptor subunits of the mTORC1 complex.34,41,75 Nevertheless, research is needed to further elucidate how amino acid signals are relayed to IPMK for its respective activation.
Significant advancements have been made in order to expand the growing body of evidence in order to bridge the gaps that presently exist in the amino acid sensing mechanisms leading to mTORC1 activation. The discoveries of various protein components that are suggested to play a role in sensing and relaying amino acid signals to mTORC1 have advanced the understanding behind the complexity of signaling cascades converging upon the mTORC1 pathway (Figure 3). The mechanisms underlying the spatial recognition and regulation of amino acid-induced mTORC1 activation are currently less characterized. Each respective finding that is proposed to be involved in amino acid signaling to mTORC1 serves a grandeur purpose in understanding the multitude of factors behind mediating mTORC1 activation. The multiple amino acid-sensing mechanisms described within this review help to present a clearer link between nutrient sensing of mTORC1, lyososomal localization components, as well as subsequent up-regulation of translation initiation for regulating MPS.
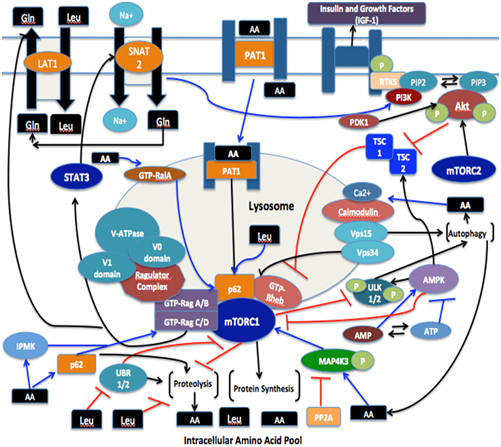
Figure 3 The various mechanistic pathways of amino acid input to mTORC1. PI3K signaling is triggered by insulin and growth factors (i.e., IGF-1), which leads to GTP-loading of Rheb. Lysosomal membrane localization of farnesylated Rheb-GTP then occurs, and upon energy depletion, this signaling cascade is negatively regulated by AMPK. The abundance of intracellular amino acids instigate GTP loading of Rags, thereby resulting in activation of Rag complexes which then transport mTORC1 to the lysosomal membranes (where it can be activated by Rheb-GTP) through interactions with the Ragulator complex and or p62. The interaction between mTOR and Raptor in response to amino acids to facilitate Rag binding is thought to be stabilized by inositol polyphosphate multi-kinase (IPMK). mTORC1 activity can be promoted by MAP4K3 and further augments by amino acids; however, PP2A functions to negatively regulate this activity in response to amino acids depletion. Amino acids are thought to regulate Ral A and positively up-regulate mTORC1 signaling. The expression of the amino acid transporter SNAT2, possibly due to activation of STAT3, is up-regulated by mTORC1. SNAT2 is Na+-linked and mediates glutamine uptake in a concentration-dependent manner, which can then be exported in exchange for leucine import by LAT1; the expression of which is also up-regulated by mTORC1. PI3K signaling to mTORC1, as well as inhibiting proteolysis, is also proposed to occur through SNAT2 activation. Proton-assisted amino acid transporter 1 (PAT1) acts as a transceptor, and is involved in transducing the amino acid signal to mTORC1 at the endomembrane surface through undefined mechanisms. Intracellular amino acid transport instigates Ca++ influx, thereby activating Vps34 which may be involved in transducing the amino acid signal to mTORC1 or increasing amino acid levels through its role in autophagy. As such, mTORC1 negatively regulates autophagy through phosphorylation ofUnc-51-like kinase 1 (ULK1), while AMPK functions to activate autophagy during a depletion of energy levels. The intracellular amino acid pool is also replenished through proteolysis, and leucine is thought to inhibit this process either directly or by inhibition of unbranched-chain amino acid receptor (UBR) proteins to promote an anabolic phenotype [34].
Continual elucidation behind each component associated with these amino acid-sensing pathways, as well as the governance of amino acid transporters in the presence of elevated amino acids, is of importance. Another area garnering additional clarification would include investigating the amino acid-sensing mechanisms through the AMPK pathway.19,34 Since it is likely that both amino acid and energy-sensing mechanisms are tightly coordinated in order to regulate MPS, exploring signaling avenues which converge upon mTORC1 is helpful. Moreover, since it is understood that both mTORC1 and Vps34 play a role in regulating the catabolic processes of autophagy, investigations into components such as the autophagy kinase,Unc-51-like kinase 1 (ULK1), may be beneficial in expanding the understanding of the amino acid-sensing mechanisms.34 In summary, amidst the significant advancements in elucidating the amino acid sensing mechanisms, continual exploration is needed to further highlight the unanswered questions behind the complexity of amino acid sensing mechanisms converging upon the mTORC1 complex.
This brief review project was not partially or fully sponsored by any external sponsoring agent, rather support was provided by the Department of Health, Human Performance, and Recreation of Baylor University, Waco, TX, USA.
Author declares that there is no conflict of interest.

©2015 Hwang, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.