Journal of
eISSN: 2473-0831


Research Article Volume 10 Issue 6
1Centre for Modern Interdisciplinary Technologies, Nicolaus Copernicus University, Wileńska 4, 87-100 Toruń, Poland
2Department of Cellular and Molecular Biology, Nicolaus Copernicus University, Lwowska 1, 87-100 Toruń, Poland
3Department of Plant Physiology and Biotechnology, Nicolaus Copernicus University, Lwowska 1, 87-100 Toruń, Poland
4Department of Pediatric Hematology and Oncology, Collegium Medicum, Nicolaus Copernicus University, M. Curie Skłodowskiej 9, 85-094 Bydgoszcz, Poland
Correspondence: Joanna Szczepanek, Centre for Modern Interdisciplinary Technologies Nicolaus Copernicus University in Torunul. Wilenska 4, 87-100 Torun, Poland
Received: October 20, 2021 | Published: December 3, 2021
Citation: Szczepanek J, Górka S, Domagalski K, et al. Whole genome and transcriptome analysis of genetic determinants associated with cyclophosphamide in vitro in pediatric acute leukemias. J Anal Pharm Res. 2021;10(6):229-238. DOI: 10.15406/japlr.2021.10.00389
Introduction: The main objective was to implement the determinations of the ex vivo resistance to cyclophosphamide and to identify the genetic profile for pediatric patients with acute leukemias.
Methods: In order to determine the ex vivo drug resistance profile, MTT cytotoxicity assay was performed on mononuclear cells. Gene expression profiles were prepared on the basis of cRNA hybridization to oligonucleotide arrays of the human genome (Affymetrix). We performed also array-based comparative genomic hybridization using a SurePrint G3 Human CGH Microarray. Data was analyzed by bioinformatics tools. Verification of the relative expression level of 20 genes was carried out by qRT- PCR.
Results: We observed a multitude of differentially expressed genes, e.g. ANXA1 (FC=3,04), BCL2A1 (FC=2,69), SERPINA1 (FC=2,12), DHRS7 (FC=2,13), PCDH9 (FC=- 4,58), TTC28 (FC=-2,25) and DUSP1 (FC=-2,91). The expression of genes that code for inflammation mediated by chemokine and cytokine signaling, Wnt, angiogenesis and integrin signaling and T cell activation pathways genes affect the sensitivity of leukemic blasts to cyclophosphamide. Transcriptome level changes are associated with chromosomal aberrations, especially located on chromosomes 8, 10, 14, 15, 16 and 22.
Conclusion: Our work delineated genes with differentiated expression and recurrent copy number changes, and revealed novel amplified loci and frequent deletions in resistant to cyclophosphamide cells, which may guide future work aimed at identifying the relevant target genes. In particular, deletion seems to be a frequent mechanism of IFIT3 gene inactivation. ANXA1, SERPINA1, TCF7 and BCL2A1 may also be included among the candidate genes of resistance (Ontological analysis).
Keywords: acute pediatric leukemia, CP, drug resistance, microarrays, comparative genomic hybridization, genetic profiling
Childhood acute leukemias (AL) are a group of diseases with varied immunophenotypes and specific genetic abnormalities.1 Acute lymphoblastic leukemia (ALL), the most common type of childhood leukemia, develops from early forms of B or T cells at different stages of maturity.2 Most of the remaining cases account that acute myeloid leukemia (AML) starts from the myeloid cells that form white blood cells (other than lymphocytes), red blood cells or platelets.3 The 5-year survival rate for children with AL has greatly increased over time and is now more than 90% overall for ALL and in the range of 60% to 70% for AML.1,4 Survival rates vary depending on the subtype of acute leukemia and other prognostic factors.1 Relapse risk can be predicted by clinical and pharmacogenetic features, early treatment response to tailor chemotherapy intensity and genetic characteristics of leukemic cells.5,6 Patients diagnosed with de novo AML and patients with secondary ALL have more resistant lymphoblasts then patients with de novo ALL. In pediatric patients with de novo and secondary AML, there is no differences in the degree of drug resistance.7 The development of better treatment for ALL and AML, and stratification of risk groups is aided by diagnosing complete cytogenetic changes, which allows for understanding of molecular abnormalities. In most patients, the associated profile of resistance to chemotherapeutics is correlated with the gene expression profile.
Cyclophosphamide (CP) is a non-cell cycle specific cytotoxic alkylating agent of the nitrogen mustard type, administered to treat malignant diseases (e.g. acute leukemias, retinoblastoma, breast and ovarian cancers). As a pro-drug, it requires metabolic activation by microsomal enzymes in order to exert activity.8 Following absorption, CP in the liver is converted into 4-hydroxyphosphamide by a cytochrome P450 isoform CYP2B.9-11 The activated form of CP, a phosphoramidate mustard, can act according to three different mechanisms: (1) alkylation of two nitrogen atoms (N7) in both strands of nuclear DNA, leading to DNA fragmentation by repair enzymes, (2) damage DNA by forming cross-links between atoms in the DNA chain, preventing separation of DNA replication from the transcription process and (3) substitution of abnormal base pairs, resulting in the generation of erroneous non-coding sequences leading to mutations. Its cytotoxic effect may be also connected with inhibition of protein synthesis, T-helper cell functions suppression and prolonged reduction of B cells.12
Understanding the mechanism of resistance to CP is unsatisfactory. It may be related to: (1) increased aldehyde dehydrogenase activity,8,13-15 (2) increased concentration of intracellular glutathione and activity of the enzyme glutathione S-transferase,16,17 (3) enhanced repair of DNA lesions,18 (4) polymorphisms of CYP2B6, CYP3A4 and CYP3A5. Many of the genetic variants that affect CP metabolism and resistance may still be unknown, therefore, there is a need for carrying a detailed analysis aimed at better understanding of the genetic basis of this phenomenon.
Patients and leukemia cells
The leukemic cells of 163 children with AL were tested for their in vitro resistance profile to cyclophosphamide (Table 1). The group included 113 de novo ALL (94 B-ALL and 19 T-ALL) and 31 de novo AML patients. In this group there were also patients with relapse of leukemia (13 ALL and 6 AML cases). The patients were divided into three groups. The expression microarray study group included 72 patients (48 de novo ALL, 8 relapsed ALL and 16 AML), for whom microarray and qRT-PCR analyses were performed. Comparative genomic hybridization was performed for 46 patients. The third group, consisting of 163 patients (including patients from arrays group), was regarded as an independent qRT-PCR validation group. This study was carried out with approval of the regional CM NCU Ethics Committee (Bydgoszcz).
|
Type of leukemia |
Expression group |
CGH group |
Validation group |
|
|
Number of patients |
N |
72 |
48 |
163 |
|
ALL |
48 |
32 |
113 |
|
|
AL relapse |
8 |
0 |
19 |
|
|
AML |
16 |
16 |
31 |
|
|
Age (median) |
N |
8 yrs |
10 yrs |
8 yrs |
|
ALL |
7 yrs |
9 yrs |
7 yrs |
|
|
AL relapse |
11.5 yrs |
0 |
9 yrs |
|
|
AML |
10 yrs |
12.5 yrs |
11 yrs |
|
|
Sex |
N |
37F:35M |
28F:20M |
81F:82M |
|
ALL |
25F:23M |
18F:14M |
58F:55M |
|
|
AL relapse |
2F:6M |
0 |
13F:6M |
|
|
AML |
10F:6M |
10F:6M |
10F:21M |
Table 1 Patients groups characteristics
Legend: N – Mount, AL – acute leukemia, M- male, F-female
In vitro resistance assay
Samples of bone marrow were collected in a heparinised tube. 4-HOO-cyclophosphamide (0.096-100 μg/ml; Asta Medica, Hamburg, Germany) was used for cellular drug resistance testing by MTT assay, as described previously.19,20 The median value of ex vivo resistance to CP in all patients was selected as the cut-off value discriminative between resistant and sensitive samples for the purpose of selecting of genes in microarray analyses. Genetic variants were compared among resistant and sensitive cells.
RNA and DNA extraction
Mononuclear cells were isolated by a Ficoll gradient, lysed in RLT buffer, frozen and stored at -80°C until the preparation for gene expression analysis. Total RNA was isolated and purified from cells using a RNeasy Mini Kit with on-column DNA digestion (Qiagen, Hilden, Germany) in accordance with the manufacturer’s instructions. Patient genomic DNA was extracted with DNeasy Blood & Tissue Kit (Qiagen) using 200 μl of lysate, according to producer recommendations. Samples were digested with RNase A (Qiagen). The quantity and concentration of the purified nucleic acids were determined by using NanoDrop ND-2000 (Thermo Fisher Scientific Inc.). RNA concentration and RNA integrity were analyzed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). The quality of the purified RNA was evaluated using Agilent 2100 Bioanalyzer with an RNA 6000 Nano Kit (Agilent). DNA purity and concentration were measured using fluorometric quantification (fluorometer Qubit 2.0; Qubit® dsDNA HS Assay Kit, Thermo Fisher Scientific).
Expression arrays
For each experimental condition 5 μg of very high quality total RNA had been provided to carry out the cRNA labeling, hybridization, and data analysis as described below.26 Preparation and purification of biotine labeled cRNA for hybridization onto GeneChip® Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA) was performed in accordance with recommended Affymetrix protocols.
Array comparative genomic hybridization
DNA was analyzed by aCGH on SurePrint G3 Human CGH Microarray, 8x60K (Agilent). gDNA was restricted, digested and labeled with the use of SureTag Labeling Kit (Agilent). Patient gDNA (0.2 μg) was labeled with Cyanine-5 CTP dye and male or female reference DNA was labeled with Cyanine-3 CTP dye (Cyanine CTP Dye Pack, Agilent). Amplified and labeled DNA was cleaned by using the reaction purification columns and TE (pH 8.0) hybridized on microarray for 24h using the procedure recommended by the producer. After hybridization, arrays were washed and scanned (SureScan Dx Microarray Scanner, Agilent).
Quantitative real time polymerase chain reaction (qRT-PCR)
To validate the array results, qRT-PCR was performed for 20 selected genes. Random hexamer primers and High Fidelity Reverse Transcriptase (Roche Diagnostics, Basel, Switzerland) were used to generate cDNA. Primers designing and selection short hydrolysis probes (from Universal ProbeLibrary Set, Human with Probes #1 to #90) were performed with the use of Roche Assay Design Center. As a reference control, housekeeping genes were used, including TUBA1B, SDHA, GAPDH and ACTB (Table 1 in supplementary materials). Real-time PCRs were performed using FastStart Universal Probe Master (Roche Diagnostics) and the LightCycler® 480 Instrument II. Messenger RNA levels were quantified using the Pfaffl method.
Analysis
Differentially expressed genes for transcriptomic profiles were identified using the significance analysis of microarrays statistics with a false discovery rate (FDR). For each gene, the correlation coefficient between gene expression level, the in vitro drug resistance (R), the p-value and the fold change (FC) were also determined. A p-value of 0.05 was considered statistically significant. Comparisons of genology (GO) and pathways were made based on lists of filtered genes for the CP resistance profile using the PANTHER classification system (Protein Analysis through Evolutionary Relationships), version 7.0 (SRI International, Menlo Park, California, USA). Data obtained after CGH arrays scanning was analyzed by bioinformatics tools: Agilent Feature Extraction Version 10.7 & CytoGenomics Edition 2.9.2.4. The analysis included: localization, type of change (deletion/gain) and size (bp). All changes were statistically significant (p-value<0.05). We used also UCSC Genome Browser on Human to visualize and browse genome with annotated data like gene and proteins identification, their expression, interaction and regulation. GeneCards was used to provide genomic, transcriptomic and functional information on all known and predicted human genes. Differentially expressed genes in qPCR were determined using the Mann-Whitney tests for the expression levels of each analyzed gene. For the comparison of qualitative data, the χ2 method exact test was used where appropriate. Logistic regression analysis with forward selection was used to identify factors that were independently associated with SVR. For all these tests, two-tailed p-values were used and p-values <0.05 were considered statistically significant. Statistical analysis and graphing were performed with IBM SPSS 20 and GraphPad Prism 6 software.
Identification and classification of differentially expressed genes
Genes were ranked by fold change (FC) in expression level as up or down regulation. Using the permuted Spearman correlation test, 144 probe sets/122 genes significantly correlated with sensitivity to cyclophosphamide in ALL (p<0.05, FC>2), were selected. Up regulation in expression was observed for 91 probe sets/80 genes. Down regulation was found for 53 probe sets/42 genes. CP chemoresistance in AML was correlated with the changes in level of expression for 152 probe sets/118 genes (57 probe sets/44 genes were up regulated; 95 probe sets/74 genes were down regulated). The largest changes in the level of expression in 10 genes for each profile are presented in Table 2.
The only gene whose expression profile changes strongly correlated (p=0.0008) with the resistance profile for both types of leukemia was PDLIM5 (PDZ and LIM domain 5, located on 4q22.3). GO annotations related to this gene include transcription factor, non-motor actin and actinin binding. A significant increase in the level of expression was observed for annexins (A1-FC=3.036; p=0.012 and A5- FC=2.722; p=0.041), which protein products are able to bind to cellular membranes in a Ca2 -dependent manner.
Gene ontology
We assigned the differentially expressed genes to biological and functional class. In the case of molecular functions most common in resistance patterns, genes performing functions related with catalytic, receptor activity and nucleic acid binding were represented. For ALL there were changes in the expression level of genes, such as: enzyme regulator activity hydrolase activity, protein binding and especially the receptor activity. In the case of AML profile genes: transferase activity hydrolase activity and DNA binding or nucleic acid binding transcription factor activity were dominated (Figure 1). The biological analysis of the ALL profile mainly concerned genes involved in cellular component organization or biogenesis, and in the case of AML, genes that function in the metabolism of the compound containing the nucleobase of the protein transport, or transcription from the RNA II polymerase promoter and the process of cell protein modification. Characteristic feature of ALL profiles was the increased expression of genes for the membrane movement protein, phosphatase signaling molecule or cytoskeletal protein Figure 1 supplementary materials. On the other hand, in AML we more often observed a decrease in the expression of gene groups belonging to the cell adhesion molecule, enzyme modulator, protease and transcription factor (Figure 2 supplementary materials).
In both in vitro resistance profiles, a number of genes that play an important role in regulating the functioning of the integrin pathway have been identified, including TSPAN7, ITGB2, FGR, RGS10 and ITGAM for ALL and TUBA1C and VCL for AML profile (Table 3). For each of these genes, except VCL (FC=2.07), at least 2-fold increase in expression level was shown. During the cytogenetic analysis, an amplification was found on chromosome 21 in the q22.13-22.3 band, where ITGB2 gene is located. β2 integrins, encoded by the ITGB2 gene, include adhesion molecules, such as LFA-1, Mac-1 and GP-150/95 glycoprotein. In both profiles, a change in the level of gene expression of Wnt pathway was observed. Decreased expression level was shown in most of the genes, i.e. PCDH9 (FC=-4.58), PRKCZ (FC=- 3.43), CTBP2 (FC=-2.66) and FZD2 (FC=-2.16), with the exception of TCF7 (FC=2.16) and a proto-oncogene MYCN (FC=2.13). Changes in the expression of these genes may increase the concentration of β-catenin, which induces gene expression, i.e. PAX5. Dysregulation of the cell cycle, pre-B cell maturation and angiogenesis can therefore lead to neoplastic transformation.
|
Pathway |
Up-regulated genes |
Down regulated genes |
CGH group |
Validation group |
|
Inflammation mediated by |
RRAS2, ITGB2, PRKACB, |
PRKCZ, MYLK |
48 |
163 |
|
chemokine and cytokine signaling |
ITGAM, PTGS2 |
32 |
113 |
|
|
Wnt signaling |
TCF7, MYCN |
CTBP2, PRKCZ, PCDH9, FZD2 |
0 |
19 |
|
Angiogenesis |
TCF7 |
EFNB2 , PRKCZ, FZD2 |
16 |
31 |
|
Gonadotropin releasing hormone |
TCF7 |
DUSP1, PRKCZ, ANXA5 |
10 yrs |
8 yrs |
|
receptor |
9 yrs |
7 yrs |
||
|
Endothelin signaling |
PRKACB, PTGS2 |
PRKCZ |
0 |
9 yrs |
|
Integrin signalling |
ITGB2 , ITGAM, ACTN1 |
- |
12.5 yrs |
11 yrs |
|
T cell activation |
CD28, CD3D |
HLA-DOA |
28F:20M |
81F:82M |
|
ALL |
25F:23M |
18F:14M |
58F:55M |
|
|
AL relapse |
2F:6M |
0 |
13F:6M |
|
|
AML |
10F:6M |
10F:6M |
10F:21M |
|
Table 3 Panther pathway analysis of differentially expressed genes in ALL (cutoff = 2 relative expression change)
CGH array analysis
In Table 4 we showed consistently amplified and deleted regions in acute leukemia’s samples. A 0.29-Mb stable deleted region involving 10 genes was detected at 10q23.31. Among the affected genes are copy number variants in the IFIT3 gene (interferon-induced protein with tetratricopeptide repeats 3), which regulates cell migration, proliferation and signaling. IFIT3 exhibits an antiproliferative activity via the up regulation of cell cycle negative regulators CDKN1A/p21 and CDKN1B/p27. Simultaneously, we have found for this gene a significant decrease of mRNA expression in resistant cells (FC=-2.15, p=0.023).
|
Chromosome name |
Set 1 = CYKL resistant patients |
Set 2 = CYKL resistant patients |
|||||||
|
Size (Mb) |
Num Gains |
Num Losses |
-log10(PVal) Gain |
-log10(PVal) Loss |
Num Gains |
Num Losses |
-log10(PVal) Gain |
-log10(PVal) Loss |
|
|
chr8 (p11.22) |
0,13 |
1 |
2 |
0,001 |
0,187 |
2 |
0 |
1,146 |
0,000 |
|
chr10 (q23.2- |
2,15 |
0 |
2 |
0,000 |
0,032 |
0 |
1 |
0,000 |
0,310 |
|
q23.31) |
|||||||||
|
chr10 (q23.31) |
0,09 |
0 |
3 |
0,000 |
0,063 |
0 |
1 |
0,000 |
0,217 |
|
chr10 (q23.31) |
0,29 |
0 |
2 |
0,000 |
0,032 |
0 |
1 |
0,000 |
0,310 |
|
chr14 (q32.33) |
0,36 |
5 |
0 |
0,032 |
0,000 |
1 |
0 |
0,310 |
0,000 |
|
chr14 (q32.33) |
0,31 |
5 |
0 |
0,028 |
0,000 |
2 |
0 |
0,398 |
0,000 |
|
chr14 (q32.33) |
0,09 |
6 |
0 |
0,046 |
0,000 |
2 |
0 |
0,301 |
0,000 |
|
chr14 (q32.33) |
0,07 |
2 |
0 |
0,032 |
0,000 |
1 |
0 |
0,310 |
0,000 |
|
chr14 (q32.33) |
0,13 |
2 |
0 |
0,032 |
0,000 |
1 |
0 |
0,310 |
0,000 |
|
chr15 (q11.2) |
1,91 |
1 |
2 |
0,011 |
0,187 |
1 |
0 |
0,456 |
0,000 |
|
chr16 (q21) |
0,11 |
1 |
2 |
0,090 |
0,032 |
0 |
1 |
0,000 |
0,310 |
|
chr22 (q11.22) |
0,10 |
2 |
0 |
0,003 |
0,000 |
2 |
0 |
0,867 |
0,000 |
Table 4 Overview of aCGH findings in resistant acute leukemias samples
A total of 12 CNAs with size 0.07 to 2.15 Mb were recognized, involving chromosomes 8, 10, 14, 15 16 and 22. The most common chromosomal change included gains of 14q32.33. In patients resistant to cyclophosphamide, an amplification was discovered on chromosome 8 in the p11.22 band, where the following metallopeptidase genes are located: ADAM metallopeptidase domain 2 (ADAM2), ADAM metallopeptidase domain 3A, pseudogene (ADAM3A), ADAM metallopeptidase domain 18 (ADAM18), ADAM metallopeptidase domain 32 (ADAM32). Functions of these genes are related to biological processes such as cell adhesion. This change may, however, be correlated rather with the pathogenesis of acute lymphoblastic leukemia, than the induction of drug resistance. One of the most frequently identified changes was the amplification in the q32.33 band on chromosome 14, where the following genes can be found: immunoglobulin heavy constant gamma 1, G1m marker (IGHG1), immunoglobulin heavy variable 3-66 (IGHV3-66) and long intergenic non-protein coding RNA 221 (LINC00221), as well as microRNA 7641-2 (MIR7641-2) and microRNA 195 (MIR195) which are involved in post-transcriptional regulation of other genes’ expression.
A frequently identified duplication was the change localized on chromosome 21 in bands q21.1-q22.3. For genes encoding ADAM family proteins, located in the band p11.22 of chromosome 8, an amplification was observed, which can contribute to significant changes in the proteolysis of proteins and cell adhesion. Moreover, among the duplicated genes there are genes encoding adhesion molecules, integrins, immunoglobulin receptors, genes involved in the proteolysis of proteins, i.e. metallopeptidase and transporters of the ABC family. Genes affecting T, B and NK lymphocyte activation, actin cytoskeletal organization and the growth, differentiation, and migration of cells, where mainly subject to deletion. In addition, we conducted another analysis which took into account the location of genes from the transcriptomic profile. On this basis, a significant over-expression of genes located on chromosomes 1, 2, 10, 12, and a decrease in the level of expression on chromosomes 20, 21 and 22 were found (Figure 3 in Supplemantary). qPCR results. This step was performed in order to verify the selected matrix genetic markers. Twenty of them were tested (Table 2 in supplementary materials), however, the greatest significance was obtained for 9 of them (Figure 2). Simultaneous decrease in expression for ABCG1 and PCDH9, and increase in expression for DUSP2, ITGAM, ANXA1, FGR, SERPINA1, HK3 and RETN were found in the CP-resistant blasts.
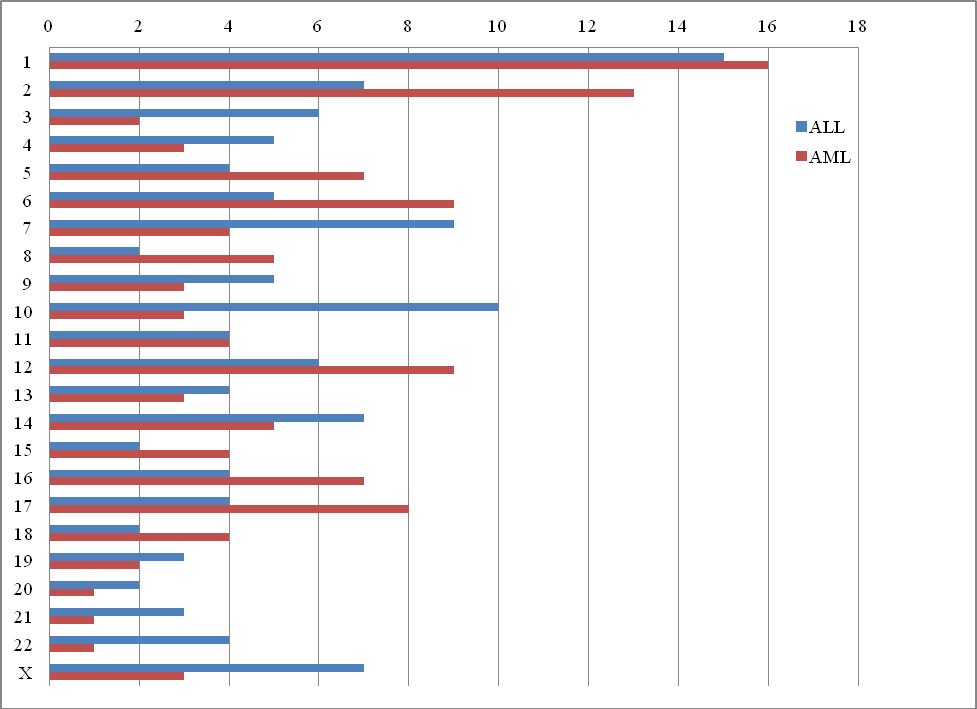
Figure S3 Distribution of all of the analyzed genes of the transcriptomic profile for ALL and AML on individual chromosomes.
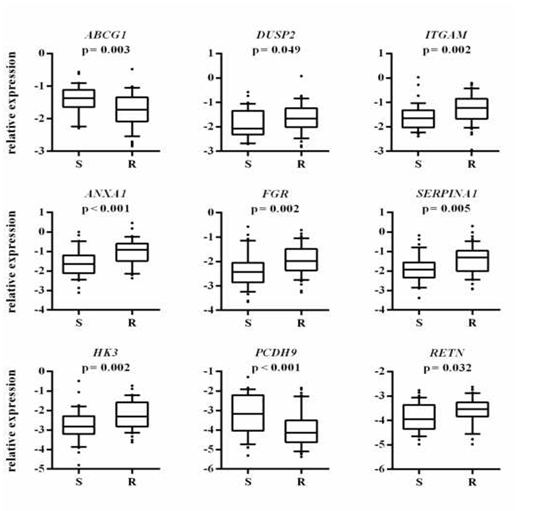
Figure 2 Association between the leukemic blasts expression of selected genes with in vitro cyclophosphamide resistance profile.
The y-axis shows the relative unit of a given gene. normalized to reference genes in log scale. as a box plot displaying the 10th. 25th. 50th. 75th. and 90th percentiles of expression levels. In vitro resistance profile is defined according to the LC50 value in MTT assay (S- sensitive. R-resistant). The P values were obtained by the Mann-Whitney test.
For the purpose of this study, transcriptomic and genomic profiles correlated with the cyclophosphamide resistance profile in the two most common types of leukemia in children, were analyzed. Among the most important mechanisms for the analyzed events, deletions and duplications were identified, located on the chromosomes: chr8 (p11.22), chr10 (q23.2- q23.31), chr14 (q32.33), chr15 (q11.2), chr16 (q21) and chr22 (q11.22). Duplicated genes include genes encoding adhesion molecules, integrins, immunoglobulin receptors, genes involved in protein proteolysis, i.e. methylpeptidases, and transporters from the ABC family. The majority of patients subjected to biological analysis had deletions. Genes influencing the activation of T, B and NK lymphocytes, organization of actin cytoskeleton as well as growth, differentiation and cell migration were subject to deletion. In ALL profile, a significant number of genes encode proteins serving as receptors: CD2, CD3, CD8A, CD22, CD28, CD99 and CD300A, and all of these genes, except CD22 (FC=-2.86), exhibit at least 2-fold increase in expression level. CD2 (FC=3.21) is a transmembrane glycoprotein whose expression occurring in T lymphocytes and NK cells allows for T-ALL diagnosis. Through interaction with other proteins, CD2 can influence on activation, proliferation and apoptosis of T lymphocytes and NK cells.21,22 Chiaretti et al., demonstrated a correlation between increased expression of CD2 in cells responsible for the relapse of T cell leukemia in adults.23 However, Uckun et al. have shown that the presence of CD2 is associated with a favorable prognosis in pediatric patients.24 On the other hand, the amplification of genes encoding proteins from the ADAM family can contribute to significant changes in protein proteolysis and cell adhesion. The phenomenon of the lack of sensitivity to cyclophosphamide may also be associated with amplification of 8p11.2 region, where ADAM2, ADAM32, ADAM3A and ADAM18 genes are encoded. Although the role of these genes is not fully understood, it is believed that they are involved in cell adhesion and intracellular signaling. Changes in the level of their expression contribute to the development of various cancers, including leukemia and breast cancer.25 Research by Koike Folqueira et al. indicates that ADAMTS1 gene (a disintegrin-like and metalloprotease (reprosylin type 2) with trombospondin type 1 motif 1) is involved in cell migration, and confirm the involvement of DUSP genes, which encode phosphatases with double activity.25 In addition, overexpression of TSPAN7 has an impact on the regulation of cell adhesion associated with cadherins and integrins.26
Changes in the organization of the cytoskeleton associated with the binding of calcium and calmodulin ions are characteristic of genes fulfilling the receptor function in the ALL profile. More than a 2-fold increase in expression levels has been demonstrated for the genes S100A12, S100A11, S100A9, S100A8, encoding signal proteins with the EF-hand motif binding calcium ions in the cytosol of monocytes and neutrophils. By mediating with TLR4 (toll like receptor 4) and RAGE receptors, they are involved in the formation of the inflammatory response and activation of NF-kB (nuclear factor kappa-light-chain-enhancer of activated B cells.27,28 Genes encoding adhesion molecules are mainly duplicated, while genes involved in the activation of T and B lymphocytes are deleted.
The AML resistance profile also found a significant overrepresentation of genes involved in signal transduction and intercellular communication. In contrast, genes involved in protein metabolism are mainly genes associated with post-translational modifications of proteins (12.82%) and proteolysis (10.25%), most of which showed a decrease in expression levels. In both profiles, most of the genes responsible for post-translational modifications were involved in protein phosphorylation and glycosylation. Changes in protein metabolism may affect the release of calcium ions from ER and cell adhesion in which integrins and cadherins are involved.
Genes encoding the cAMP-dependent A kinase (AKAP2, AKAP7, AKAP10) have an increased expression level (except AKAP10), and play an important role in the ALL in vitro resistance profile. Phosphatase (PTPRC, PPP1R12A, PTPN11), most of which have a decreased level of expression, play an important role in expression profile of AML patients. The deletion of PTPRU, PTPRT genes encoding the phosphatase was determined by cytogenetic analysis, which may confirm the decrease in expression of genes encoding protein phosphatase observed in a transcriptomic study.
In this study, in some of the patients an amplification was found in the q21.13-q22.3 region on chromosome 21, where ABCG1 genes belonging to the G subfamily are located. Resistance of tumor cells to chemotherapeutics by this group of proteins may refer mainly to the changes in drug transport between the extracellular space and the interior of the cell, and the cell organelles’ membranes, to the metabolism of the drug and to the expression of enzymes responsible for the uptake of the drug molecules.29 In these transcriptomic profiles in patients with ALL and AML, there has been no change in the expression levels of genes serving as ABC transporters. The amplification of the q21.1 band of chromosome 20 was found by Itzhar et al., in patients with chemotherapy-induced AML.30
In addition, changes in the expression of the genes responsible for regulating the transcription under the RNA II polymerase promoter may be associated with abnormal expression of other genes involved in the regulation of cell division and induction of cytochrome P450 activity. Changes in the activity of P450 cytochrome isoforms (2B6, 2C9, 2C19, 3A4) aldehyde dehydrogenase (ALDH), aldo-keto reductase (AKR1) and glutathione transferase (GST), and the presence of their activators, inhibitors, and genetic polymorphism may affect the emergence of drug resistance. During the cytogenetic analysis a deletion of q23.1-q32 on chromosome 5, where ALDH7A1 gene is located, was observed. The amount of therapeutically active metabolites may affect the change of expression of the above genes, especially its growth.31 The analyzed transcriptomic profiles of ALL in vitro resistance demonstrated an over-expression of DHRS7 (dehydrogenase/reductase (SDR family) member 7) and SERPINA1, located on chromosome 14, which are involved in the metabolism of the steroid compounds, such as hormones, prostaglandins, lipids and xenobiotics.30 In the ALL expression profile an almost 3-fold increase in expression level for ANXA1 and ANXA5, which encode annexins binding calcium ions and phospholipids, and are involved in signal transduction, cell growth and differentiation, was observed.32 A change in the expression level in the ALL profile was observed for the TTC28 gene (FC=-2.33), which belongs to transmembrane proteins involved in guanine nucleotide exchange and cytokinesis.
Overexpression of anti-apoptotic proteins and a decrease in expression levels for the genes responsible for the induction of cancer cell apoptosis were observed in both profiles. Based on the analysis of the in vitro resistance profile, a decrease in the expression level of PAX5 gene (FC=-3.29) in ALL and MLL3 gene (FC=-2.79) in AML was shown. The deletion of p21.3-p13.1 and p24.3-p21.3 bands, on which these genes are located, may be associated with decreased levels of their expression. Studies by Liu et al., indicate that the loss of a PAX5 gene may affect the value of DFS (disease free survival) in children with B-ALL, by participating in the maturation and proliferation of B lymphocytes.33 MLL is one of the transcription coactivators and plays an important role in early hematopoiesis. The consequence of fusion of this gene with other genes, is the production of new chimeric proteins.34 In addition, the SOX11 gene (FC=-11.69) has two functional domains as an activator or repressor of other genes, e.g. PAX5.35,36 In AML resistance profile, the FOXC1 (FC=-99.24) and HOXA11 (FC=-15.76) genes may have a negative effect on the regulation of proliferation in several tumor cell lines.37 On chromosome 20, in patients cells sensitive to cyclophosphamide, a deletion of the q11.21-q13.33 band, where TP53INP2 and BCL2L1 genes are located, was identified. TP53 mutations are present in nearly 50% of all types of cancers and 30% of breast cancers (>15 000 different mutations).38,39 TP53INP2 gene encodes nuclear protein which is involved in the response of cells to stress, DNA damage, hypoxia, immune response and transcription regulation of other genes, e.g. Ras oncogenes.38-40 BCL2A1 (FC=2.69 in the ALL profile) and BCL2L1 genes, involved in the regulation of apoptosis, have an anti-apoptotic effect. The over-expression of BCL2A1 gene and MYCN oncogene (FC=2.13 in the ALL profile) may be associated with abnormal signal pathway of programmed cell death dependent on TP53.41 Viability of tumor cells may depend on the relationship of appropriate amounts of pro- and anti-apoptotic proteins. VDAC3 gene (FC=2.16 in the AML profile), participates in the flow of cytochrome c by encoding an inner mitochondrial membrane voltage-dependent anion channel (VDAC). Over- expression of BCL2L1 and VDAC3 can contribute to the closure of VDAC’s diameter and the inhibition of apoptosis. Overexpression of anti-apoptotic proteins and a decrease in expression levels for the genes responsible for the induction of cancer cell apoptosis were observed in both profiles.
The drug resistance phenomenon involves various groups of genes, encoding primarily transcription factors, proteins (pro- and anti-apoptotic) regulating the process of leukemia cell apoptosis, receptor proteins, hydrolases, transferases, enzyme regulators and genes responsible for the metabolism of anti-cancer drugs. Based on research, it can be concluded that cyclophosphamide influences primarily the regulation of the expression of genes which function as receptors of immune cells involved in the activation of T and B lymphocytes, and NK cells, cell adhesion (FOCAD, ACTN1, DIAPH2, COTL1, EFNB2, FGR, GNE, PCDH9, FZD2, RGS10), cell division, MAPK pathway (PRKCZ, BMX, VPREB3, DUSP1, MAP3K6), DNA replication (DHX9, S100A12, S100A11, S100A9, S100A8, S100P), mainly located on chromosome 1, and genes responsible for autoimmune diseases (inflammation). To sum up the transcriptomic data, the mechanism of action of cyclophosphamide is similar in both types of leukemia. However, there are varied groups of genes whose changes in genetic profile correlate with in vitro resistance to cyclophosphamide. Cyclophosphamide resistance in children with ALL and AML is closely correlated with changes in gene expression profiles, cytogenetic changes and, as a consequence, the coexistence of many mechanisms. In both profiles, the cytostatic resistance mechanism mainly involves changes in the metabolism of nucleic acids, mainly RNA. The effects of cyclophosphamide on cell pathways are different in both leukemias due to the cell line that prevails in them. Disruption of the apoptosis process is a common feature for the analyzed profiles. Many clinical studies on identifying genetic determinants of cyclophosphamide sensitivity indicate that the effectiveness of cancer treatment depends on proper identification of the cellular target for chemotherapeutics. However, the identification of a universal multidrug resistance gene in various tumor types, as indicated by numerous research, is currently impossible.
This study was supported by Grant from the National Science Centre No. DEC- 2011/03/D/NZ5/05749.
The authors declare that there are no conflicts of interest.

©2021 Szczepanek, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.