MOJ
eISSN: 2374-6912


Editorial Volume 3 Issue 2
Center for Genetic Medicine, Northwestern University, USA
Correspondence: Sasha Bogdanovich, Center for Genetic Medicine, Northwestern University, 303 E. Superior St. Chicago, IL 60611, USA
Received: January 29, 2016 | Published: February 10, 2016
Citation: Bogdanovich S. Gene editing with CRISPR/Cas9: a new therapeutic hope for duchenne muscular dystrophy. MOJ Cell Sci Rep. 2016;3(2):42-44. DOI: 10.15406/mojcsr.2016.03.00050
Duchenne muscular dystrophy (DMD) is a devastating disease resulting in progressive muscular weakness throughout life leading to death. This disease affects 1 in 3500 boys, who will die from cardiopulmonary failure in their early adulthood. Treatment options have been limited to palliative approaches. CRISPR/Cas9, a novel gene therapy has recently showed a great potential in animal trials to correct the primary genetic lesion of DMD. These recent studies have put into question whether such trials should be translated into higher mammalian or even human studies.
In 1861, a French physician, Guillaume Duchenne, first described DMD as “pseudohypertrophic muscle paralysis” that preferentially afflicts males within families”. It is known as one of the most common X-linked diseases, DMD affects 1 in 3500 male newborns and is the result of mutations in the dystrophin gene. Dystrophin is one of the largest known genes in the human genome, containing 79exons and yielding a 14kb transcript.1 Dystrophin protein links the extra- and intra-cellular cytoskeleton, neutralizing stressful events that come from outside the cell to intracellular matrix (Figure 1). When there is a lack of dystrophin protein, there is lack of control for the stress and cell damage occurs.2 Disease symptoms can vary from mild to severe, depending on the mutation in dystrophin gene. In the case of nonsense mutations, there is a premature stop codon that completely blocks dystrophin translation, resulting in lack of dystrophin and severe clinical presentation. In frame shift mutations, there is an exchange of one nucleotide base with another. Compared to the nonsense mutation, the frame shift mutation of the gene results in a milder clinical presentations. When muscles are biopsied, patients who have milder symptoms typically have less functional or truncated dystrophin. This milder form of disease is often referred to as Becker muscular dystrophy.3
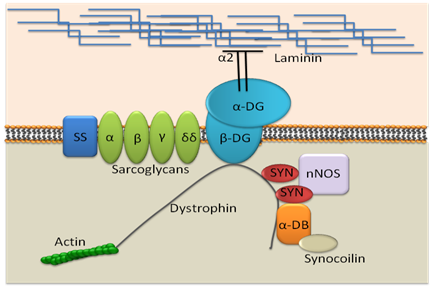
Figure 1 Dystrophin and associated protein complex at cell membrane.
SS: Sarcoglycan/Sarcospan complex; NOS: Neuronal type nitric Oxide Synthase; Syn: Syntrophin; DB: Dystrobrevin.
DMD is characterized as a disease that results in progressive muscle weakness and muscle degeneration. DMD manifests at an early age of 3 to 5years, beginning as weakness in the upper and lower limbs.4,5 Clinically, this can be identified by difficulty with rising from the floor in a stereotyped fashion known as the Gowers’ sign.6 Muscle weakness continues throughout life as cycles of degeneration, and regeneration eventually fails. When vital muscles can no longer regenerate, patients succumb to their disease, ultimately dying of cardiopulmonary insufficiency.7,8 As respiratory muscles fail later in life, mechanical ventilation is added to therapy along with other drugs aimed at reducing the progression of cardiomyopathy and respiratory infection.9,10 Patients with DMD die in their second or third decade of life, typically from respiratory or cardiac failure. Unfortunately, currently there is no medication targeted to treat this deadly disease and correct the primary lesion. Treatment is palliative and focused around reducing muscle inflammation and pain with corticosteroid therapy.11,12
Until recently, research into possible cures for DMD have focused around two methodologies:
Change of endogenous dystrophin:
Restoration of a proper reading frame to endogenous dystrophin; mRNA by using anti-sense RNA oligonucleotides to bridge afflicted exons and translate a shorter, but still functional form of dystrophin.13 Interfering RNA oligonucleotides aim to skip lesion-containing exons during translation, restoring expression of a truncated but partially functional dystrophin protein. Although successful in animal trials, this therapy is still considered premature, which has prevented its Food and Drug Administration approval.14 Another approach uses chimeroplastic injection of chimeric RNA or DNA. Both have had some success but have many limitations as well.15,16
Delivery of non-endogenous dystrophin protein via viral vectors:
Dystrophin’s large size makes it virtually impossible to package into most viral capsids.17 Therefore, shortened mini proteins (“e.g. mini dystrophin”) that can fit in common viral vectors are currently being examined and have shown promising results. The dystrophin protein consists of 3645 amino acids with molecular weight of 426kDa. It is a long molecule and can be divided into 4 parts: an N-terminal actin binding domain, a central rod, a cysteine-rich segment, and a C-terminal end (Figure 2). Mutation can occur in any of these four parts. The cysteine-rich domain has multiple functions and mutations in this part are often the most severe. Mutations in the other three portions of dystrophin protein typically result in milder clinical presentation.18,19 Mini-dystrophinwhich is able to be packaged in viral vectors typically does not contain the less important rod domain of dystrophin.20,21 In this way, the protein can maintain its function, as the other parts of dystrophinare functionally more important in preventing the disease.18,19
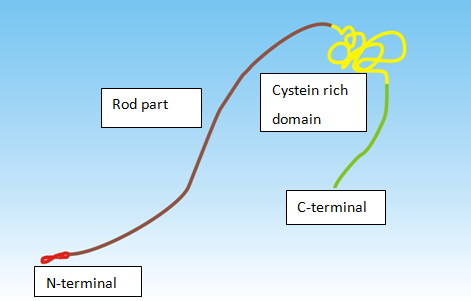
Figure 2 Dystrophin molecule. Parts of dystrophin molecule:
CRISPR/Cas9 based technology illustrates a new promise of gene therapy capable of restoring endogenous dystrophin expression without the use of oligonucleotides or recombinant dystrophin.22 CRISPR-based technology works similarly to oligonucleotide therapy in that it restores expression of endogenous dystrophin. Unlike oligonucleotide therapy, CRISPR based technology removes the affected exon from the host genome altogether, not requiring indefinite treatment like pharmacological therapies.11,12 The CRISPR/Cas9 system evolved in bacteria as a host defense against foreign genomic material, enabling the targeted deletion of specific sequences.23 Recently, this system has been redesigned to remove lesions from many mammalian genomes in the hope of one day correcting a range of human genetic diseases.24 This technology has already been used to restore dystrophin expression in tissues of the mdx mouse, an animal model of DMD. These data were recently described in journal of Science by three independent research groups from Duke University, University of Texas Southwestern Medical Center, and Harvard University.22,23,25 Nelson et al.,25 has recently made particularly remarkable advances in remodeling CRISPR-based technology to treat DMD. In their study, they used Adeno-associated virus to deliver the CRISPR/Cas9 gene editing system to delete exon 23, the disease causing exon in the mdx mouse. Dystrophin expression was restored in these mice, and ameliorated the dystrophic phenotype altogether. The major hallmark of the study was the systemic, intravenous delivery of their gene editing complex and dystrophin restoration in many different muscles, including the heart, which had proven to be exceptionally difficult to target in the past.25
Although very promising, CRISPR-based technology requires further research before translation to human studies. How the CRISPR/Cas9 gene editing system will react with the human immune system is still entirely unknown. Another potential concern is off-target cuts, or the erroneous excision of genetic material; this is arguably the greatest disadvantage of this system. In addition, a potential therapeutic approach that seeks to explore gene editing in humans will likely raise many ethical questions. However, even though there are many questions to be answered, CRISPR/Cas9 gene editing system is definitely the most successful gene therapy to date used in an animal model of DMD, and it will be exciting to see how this furthers the treatment of the disease in humans.
None.
The author declares no conflict of interest.

©2016 Bogdanovich. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.