Journal of
eISSN: 2378-3184


Review Article Volume 7 Issue 1
1Aquaculture Faculty of Animal Husbandry Fisheries University of Tadulako Indonesia
2Microbiology and Immunology School of Veterinary and Biomedical Sciences James Cook University Australia
Correspondence:
Received: September 13, 2017 | Published: January 12, 2018
Citation: Rusaini, Owens L (2018) Suppression Subtractive Hybridization in Penaeid Prawns: An Approach in Identifying Diseases and Differentially Expressed Genes. J Aquac Mar Biol 7(1): 00177 DOI: 10.15406/jamb.2018.07.00177
The polymerase chain reaction (PCR)-based cDNA subtraction method called suppression subtractive hybridization (SSH) is one of the most powerful methods in identifying differential gene expression in a given sample. The SSH technique offers many advantages in profiling gene transcripts. It enriches the sequence of interest over 1000-fold in a single round of hybridization, allows for the representation of low abundance genes in the library and prevents undesirable cDNA amplification. This subtractive PCR technique has been used to study the expression of genes in penaeid prawns challenged with viruses, bacteria, and environmental stressors. Many functionally important genes of penaeid species have been identified from the SSH cDNA libraries. These differentially expressed genes were clustered based on the sequences having significant homology in the GenBank database. However, the regulatory mechanism and the exact role of these genes in the immunity of penaeid prawns are mostly unknown.
Keywords: suppression subtractive hybridization, differentially expressed genes, penaeid prawns, viruses, bacteria, environmental stressors
Alteration in gene transcripts is related to a wide spectrum of physiological and pathological changes in cells. Therefore, the identification of differential gene expression can be used to understand biological processes at a molecular level.1–4 Various techniques such as Expressed Sequence Tags (EST), differential display, Serial Analysis of Gene Expression (SAGE), cDNA microarray, and subtractive hybridization have been developed to analyse differential gene expressions in any given sample.5,6 The majority of methods used for differentially expressed genes are based on differential display and subtractive hybridization. As it is technically demanding and labour intensive due to the need for library screening to identify isolated genes, subtractive hybridization is less widely used than differential display.6,7 However, subtractive hybridization produces fewer false positives compared to differential display.5,6
In penaeid prawns, differentially expressed genes, in particular immune-related genes are rarely investigated. In fact, this information is important in understanding host-pathogen interactions at the molecular level during infections. It may provide a solution to maintain prawn production since many previous efforts seem to be unable to circumvent the catastrophic effect of infectious diseases in prawn aquaculture.8 The ability of the SSH technique to discover many functionally important genes in a single sample offers a great opportunity in understanding differential gene expression in penaeid prawns. Several aspects of the subtractive hybridization technique, including its application in various biological questions and its comparison with other related methods for differentially expressed genes, have been reviewed.5,6,9 This review will draw particular attention to suppression subtractive hybridization as a method to profile differential gene expression and its application in elucidating diseases and host immune-genes of penaeid prawns.
Early development of suppression subtractive hybridization
Initially, subtractive hybridization was used to study different expression of genes in the developmental stage of Xenopus laevis10 and in T-helper hybridoma cell lines.7 Modifications of this method have been applied to a cDNA library in scrapie-infected brain of the hamster.11,12 Commonly, this technique includes cDNA hybridization from one population to excess cDNA from another population and separation of unhybridised fragments (target) from hybridised common fractions.13 Mostly, the common sequences in the two populations were eliminated by hydroxylapatite chromatography10,7 an avidin-biotin system11,12 or oligo(dT)30-latex beads.14,15 Later, integration of PCR into subtractive hybridization increased the sensitivity of the subtraction method and led to the possibility of using low abundance Mrna.16 This integrated method has been used in identifying differentially expressed genes in thyroid hormone-induced tadpole tail regression,17 in the human placenta of a newborn female and in Epstein-Barr virus-immortalised lymphoid cell lines,18 human embryonal carcinoma cells,14 and human diploid fibroblasts.15
However, to some extent, the use of subtractive hybridization is limited by its complexity due to the nature of subtraction kinetics. Low abundance genes may hybridize slower and obtain a lower level of completion compared to high abundance genes. Thus, it works poorly in identifying low abundance genes. Unfortunately, many target genes involved in cellular processes are in low abundance, so to maximise the identification of rare messages, high concentration of primers are necessary at the expense of insert length. In addition, several rounds of hybridizations may be required to prevent retention of sequences that are not differentially expressed.5–19 However, the number of common cDNA clones still remained high in the two samples.15
To overcome these problems, recently subtractive hybridization has been modified or combined with other differential gene expression methods leading to the establishment of a number of related methods. These techniques include Phagemid Subtractive Hybridization, Subtractive Hybridization and Shot-Gun Sequencing, Direct Random Oligonucleotide Prime (DROP) Subtractive Hybridization, Chemical Cross-Linking Subtraction (CCLS), and Representational Difference Analysis (RDA). The modification of subtractive hybridization has also resulted in several other methods such as Combined Subtractive and Display Techniques, Subtractive Differential Display, Differential Subtractive Hybridization, Gene Profiling Using Subtractive Hybridization, and Suppression Subtractive Hybridization (SSH).6 Indeed, every method has certain advantages and disadvantages and careful consideration is needed to choose a suitable method for a specific sample, because not all methods are well-matched with all samples.
Principle of suppression subtractive hybridization
The principle of Suppression Subtractive Hybridization (SSH) is based on cDNA sequences of two samples being compared by hybridization. These two different cDNA populations are referred as “tester” and “driver”.18 Tester contains specific target (differentially expressed) sequences that are not present in driver, the reference cDNA. The common sequences of both samples are subtracted leaving a cDNA population enriched for sequence preferentially expressed in the tester but absent in the driver.13,16,19 Thus in the SSH method, target cDNA is selectively amplified, while an undesirable sequence is simultaneously suppressed during PCR amplification. The suppression PCR effect is mediated by long inverted terminal repeats attached to the end of the cDNA fragments. The inverted repeats form stable panhandle-like loop structures following denaturation and annealing procedures. This panhandle-like structure cannot be exponentially amplified in a PCR with primers obtained from long inverted repeats sequences, because intramolecular annealing of the long inverted terminal repeats is highly favoured and is more stable than intermolecular annealing of shorter PCR primers.13,19
In general, SSH involves several steps (Figure 1). Firstly, isolation of total RNA or poly (A)+ RNA is carried out from tissues or cells being compared. The tester and driver double stranded (ds) cDNA are synthesised separately from mRNAs and digested with a four-base cutting restriction enzyme (Rsa I) to obtain blunt ended cDNAs. Following this, tester cDNA is divided into two portions and each sample ligated with different adaptors (adapter 1 and adapter 2R) to the 5' end of cDNA, creating two populations of tester. The adaptor’s end has no phosphate group. Therefore, only the longer strand of each adaptor can covalently attach to 5' ends of the cDNAs. Next, the driver cDNA is mixed with each sample of testers, heat denatured and allowed to anneal, generating type a, b, c and d fractions in each sample. During this first hybridization, normalisation of the single stranded (ss) cDNA tester fragment (a) occurs, meaning the concentration of high and low abundance cDNAs become approximately equal. This equalisation occurs as a result of the annealing process generating homohybrid (b) and heterohybrid (c) fragments more rapidly for more abundant fractions due to the second-order kinetics of hybridization. Annealing of low abundance cDNAs is slower and these remain single stranded (a). Simultaneously, the ss cDNA tester molecules (a) are enriched significantly for differential gene expression while common non-target fragments form type c fractions with the cDNA driver.13,19
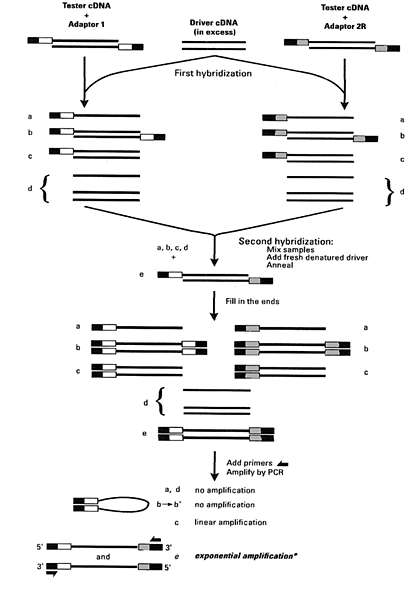
Figure 1 Schematic diagram of suppression subtractive hybridization (SSH) technique. The two different cDNA populations are synthesised from mRNA generated from tissues or cells being compared and refer as “tester” and “driver”. Tester contains specific sequence being targets that are not present in driver, the reference cDNA. Solid lines symbolize digestion of tester and driver cDNAs with a four-base cutting restriction enzyme (Rsa I). Solid boxes characterise the outer part of adaptor 1 and adaptor 2R that associated to PCR primer 1 sequencing. Clear boxes represent the inner part of adaptor 1 that corresponds to nested PCR primer 1 sequencing. Grey boxes symbolize the inner part of adaptor 2R that associate with nested PCR primer 2R sequencing. Type e fractions can only be formed if there is up-regulation of transcripts in the tester cDNA. From Clontech Laboratories, I. PCR-SelectTM cDNA Subtraction Kit User Manual, 44pp. Clontech Laboratories, Inc., Mountain View, California (Clontech Laboratories 2008).
In the next step, both samples from the first hybridization are mixed and annealed with fresh denatured driver to enrich differentially expressed gene fractions. During this second hybridization, only the remaining normalised and subtracted ss tester cDNAs can re associate and form a new type of hybrids (e). However, the type e molecules can only be formed if the transcript is over expressed in the tester cDNA. Addition of freshly denatured driver cDNA increases the extent of hybridization, causing further enrichment of type e fractions for differentially expressed sequences. These new hybrids (e) have a different feature from the other fragments in that they have two different adaptor sequences in their 5' ends, one from sample 1 (adaptor 1) and the other from sample 2 (adaptor 2R). Prior to the initiation of PCR amplification, an extension reaction is conducted to fill in the adaptor ends by DNA polymerase. This process provides a primer binding site for PCR amplification. Several types of molecules are formed with diverse combinations of adaptor sequences at their ends. Type e hybrids have distinct annealing sites for nested primer on their 5' and 3' ends.13,19,20
Finally, the mixture is subjected to two rounds of PCR to exponentially amplify the target differentially expressed genes. During the primary PCR, type a and d fractions cannot be amplified because they have no primer binding sites. Due to the suppression PCR effect, type b fragments are also unsuitable for amplification because these molecules have long inverted repeats on the ends and form a stable panhandle-like structure following the denaturation and annealing cycles (Figure 2). Type c molecules can only be amplified linearly because they only contain one primer annealing sites. Only type e molecules – the differentially expressed sequences – that have different adaptor sequences on the ends can be exponentially amplified by PCR. In the secondary PCR amplification, the differentially expressed sequences are further enriched and any background PCR products are reduced.13,19,20 The PCR products can then be inserted to a cloning vector to construct the cDNA libraries.

Figure 2 The suppression PCR effect is mediated by long inverted terminal repeats attached to the end of cDNA fragment. The inverted repeats form stable panhandle-like loop structure following denaturation and annealing procedures. This panhandle-like structure cannot be exponentially amplified in a PCR with primers obtained from long inverted repeats sequences, because intramolecular annealing of the long inverted terminal repeats is highly favoured and is more stable than intermolecular annealing of shorter PCR primers. From Ghorbel MT & Murphy D4 Suppression Subtractive Hybridization. In: Neuropeptides: Methods and Protocols (ed by Merighi A), pp. 237-259.
Advantages and disadvantages of suppression subtractive hybridization
Construction of a cDNA library using suppression subtractive hybridization is one of the most powerful approaches in identifying differentially expressed genes in a given sample.15,21,22 This technique offers many advantages in the study of the profile of gene transcripts. It enriches the sequence of interest over 1000-fold in a single round of hybridization, allows the representation of very rare messages in the library because high and low abundance sequences are equalised, and prevents undesirable cDNA amplification.1,3,11,18,13,19,23,24
Suppression subtractive hybridization also appears to be more discriminating and allows comparison of differentially expressed genes from two different isolates.25 This technique can be used to define gene alteration, both up-regulated and down-regulated genes.17,26 The up-regulated transcripts can be detected if the cDNA containing the specific target (differentially expressed) sequences is used as a tester, while the reference cDNA is used as a driver and this is termed forward subtraction. Conversely, the down-regulated genes can be identified if the reference cDNA serves as the tester, while the specific target cDNA serves as the driver (reverse subtraction) (Clontech Laboratories 2008).20 The ability of this technique to identify alteration of genes has been confirmed using independent protocols such as real-time PCR27,24 and real-time quantitative (q)RT-PCR.28–30
Another advantage of this PCR-based cDNA subtraction method is that recombinants can be amplified by a single oligonucleotide primer in the PCR. In addition, a wider representation of sequences can be produced because of the efficiency of bacterial transformation.12 This technique makes it possible to simultaneously identify expression of many genes.23 The diversity of genes found in the SSH libraries offers an opportunity to discover functionally important genes.30 Furthermore, subtractive hybridization libraries can be used continuously to carry out different experiments.11
Despite its benefit, the subtractive method also has some disadvantages. These drawbacks include: under the standard procedure, commonly 2-4µg of poly (A) RNA are needed as a starting material to construct both forward and reverse libraries. To some extent this quantity of poly (A) RNA maybe difficult to obtain. The abundance of mRNA of a differentially expressed gene should account for at least 0.01% of the total mRNA in order to be identified, because the completion of hybridization in the second hybridization step cannot be achieved if the target cDNA is too low. However, the large amount of mRNA does not accurately reflect the amount of encoded protein. In addition, genes that may have an important function in a biological process may not be detected if their mRNA abundance is not significantly altered by the event in question. In order to be expressed in the SSH libraries, the amount of target cDNA should be concentrated fivefold or more in the tester than in the driver. Very low abundance genes have the higher probability of being over expressed due to the lack of a site for the restriction enzyme. Furthermore, genes with limited poly(A) tail may also not be detected by this method. At least the genes in question should have four or more poly(A) tail in order to be detected using this PCR-subtraction method because the cDNA synthetic primer of this technique contain four poly(T) at the first 5΄ end. Low efficiency and high number of false positive clones may occur due to annealing with nonspecific PCR primers, non-specific annealing of suppression adaptors during subtractive hybridization, or incomplete subtraction of particular genes in the library.2,17,15,23,28,23,24
The four-base cutting restriction enzyme (Rsa I) used to produce small fragments (~ 600bp) in order to increase the efficiency of SSH technique, may be a disadvantage if a full length differentially expressed cDNA is required. However, creating each cDNA into multiple fragments has advantages. A complex network may result from long cDNA fragments preventing the formation of appropriate hybrids which are needed to bind the two adapters (adaptor 1 and adaptor 2R) at the end of the target sequences. In addition, small cDNA fragments produce a better representation of individual genes. Furthermore, the average small size (~200bp) of inserts in the cDNA libraries is another problem of the SSH method. This is due to the fact that short fragments are hybridized, amplified and cloned more efficiently than long fragments.13,19
To circumvent the limitations and increase the efficiency in determining differential gene transcripts, suppression subtractive hybridization has also been integrated with other techniques. Different components of several methods including differential display, SAGE, EST and SSH have been integrated into a linear system for gene identification. This integrated procedure results in reduction of sequence redundancy, a decrease in the requirement of mRNA, an increase in the accuracy of subtraction efficiency, and an increase in the likelihood of EST database matches and novel sequence identification.31 Combination SSH with subtractive hybridization and differential screening has been performed to ensure the expression of rare messages, eliminate undesirable common transcripts, and reduce the probability of false positives.32 SSH has also been combined with cDNA array hybridization,1 mirror orientation selection (MOS)23,33 and microarray analysis26 to eliminate the proportion of background clones that represent non-differentiated genes in the libraries. In addition, two different approaches of SSH called Direct Forward Subtractive Library (DFSL) and Enriched Forward Mirror Orientation Selection (EFMOSI) have been performed. It was found that DFSL was less labour intensive, technically less demanding, and had higher diversity in gene hits than EFMOSI.2 Furthermore, SSH has been modified to permit the identification of differential viral and cellular genes expressions from two cDNA populations.35
Application of suppression subtractive hybridization in penaeid prawn gene studies
Suppression subtractive hybridization as a single method for gene expression has been applied in a large number of circumstances. However, this review will focus on aquaculture studies with respect to prawn genes and diseases. The SSH method has been used to determine genes involved in growth36–39 and reproductive maturation,30 of penaeid prawns. Differentially expressed genes in the lymphoid organ of penaeid prawns with spehoid cells have been identified using this SSH method.40 This PCR–based cDNA subtraction method has been performed to elucidate the expression of immune–related genes in response to white spot syndrome virus (WSSV)41–44 and yellow head virus (YHV)45,46 infections in penaeid prawns. Some genes that encode proteins involved in the immune response of penaeid prawns to heat–killed microorganism challenges including the fungus Pichia pastoris, Gram–positive bacteria Bacillus subtilise and Gram–negative bacteria Escherichia coli have been identified using the SSH technique.21 Subtracted cDNA libraries have also been constructed to identify genes of penaeids that are expressed differentially upon challenge with Vibrio penaeicida, V. harveyi27,47,48 and lipopolysaccharide (LPS), a predominant component of Gram–negative bacterial cell–walls.49 In addition, differential gene expression in prawns due to environmental challenge including osmotic, hypoxic and hyperthermic stresses has been studied using the SSH method.28,50
Combination of suppression subtractive hybridization with other techniques has been applied to determine the differential gene expression profile of penaeid prawns challenged by virus, bacteria and environmental stress. The SSH combined with EST,51 differential hybridization,22,24 mirror orientation selection52 or microarray analysis53–55 has been used to elucidate genes involved in the immune response of penaeid prawns during WSSV infection. The SSH coupled with microarray analysis has also been applied to isolate differentially expressed genes in prawns challenged with heat–killed microorganisms and dsRNA injection55 and to identify and characterise genetic markers of V. nigripulchritudo virulence in prawn.25 In addition, the integration of these two methods has been used to study the alteration of gene expression of penaeids due to environmental stress such as hypoxic, hypoosmotic and hyperthermic conditions.29
Differential gene expression from SSH library of penaeid prawns
Several species of penaeid prawns including the black tiger prawn P. monodon, the kuruma prawn P. Japonicus,42 the Pacific blue prawn P. Stylirostris,27 the fleshy prawn P. chinensis54,39 the banana prawn P. merguiensis56,37,40 the Indian white prawn P. indicus44,48 and the giant freshwater prawn Macrobrachium rosenbergii57,49 have been used to construct the cDNA libraries for differentially expressed genes. Most of these libraries were constructed from the haemocytes, the hepatopancreas and the gills. The SSH libraries have also been constructed from cephalothorax, testis, ovaries, optic lobes of eyestalks36 and whole postlarvae.47,48
Many gene encoding proteins have been identified in the SSH cDNA libraries of penaeid prawns. These genes were clustered according to their predicted functionalities. For an instance, in WSSV infected prawns, differentially expressed genes were classified belonging to defence–related proteins, defence–related enzymes, apoptotic–related proteins, antioxidant enzymes, regulation and signal transduction factors.42–44 In general, these differentially expressed genes were clustered based on the sequences having significant homology in the GenBank database (National Centre for Biotechnology Information). However, the regulatory mechanism and the exact role of these genes in the prawns’ immune response are poorly understood and needs further investigation.
Alteration in gene expressions from SSH cDNA libraries of penaeids has been reported in a number of articles (Appendix S1). Among those, genes encoding proteins for haemocyanin, transglutaminase (TGase), lysozyme, penaeidins, thioredoxin reductase, trypsin, and elongation factors were found to be up–regulated in prawns due to viral, bacterial and environmental challenges suggesting the important role of these genes in the defensive response of penaeid prawns. Over expression of genes that encode viral proteins, has also been identified in the cDNA libraries of penaeids experimentally infected with WSSV and YHV.24,46 Conversely, genes coding for C–type lectin and cathepsin were down–regulated in prawns challenged with viruses and environmental stressors.
A problem with the study of altered genes from SSH cDNA libraries is the inconsistency of gene alterations reported by different authors and also within the same papers. Some of these genes were reported to be undergoing both up–regulation and down–regulation following different or the same treatments. For example, the gene coding for an antioxidant glutathione S–transferase was reported as being overexpressed in the hepatopancreas of P. japonicus following WSSV infection.42 In contrast, this gene was suppressed in the hepatopancreas of P. vannamei upon WSSV infection.55 An antimicrobial peptide lysozyme was up–regulated following WSSV infection at 32°C, but it was down regulated upon dsRNA treatment in the haemocytes of P. vannamei55 perhaps due to the interfering RNA phenomenon. The most worrying example on the discrepancy of gene alteration in which genes encoding for tachylectin–5A and serine protease inhibitors (serpins) experienced alteration in both directions was in the haemocytes of P. vannamei following WSSV infection at 32°C without any explanation on the phenomenon from the authors. Therefore, many more attempts in improving the quality and quantity of immune–related gene studies are needed before the suppression subtractive hybridization data gains its full potential as a technique to provide better knowledge about the immune system of the penaeid prawns.
The biggest challenge faced by the prawn aquaculture industry worldwide is to control the devastating impact of infectious pathogens. Therefore, many efforts have been made to control this problem. To some extent good management practices are effective, but other protection methods are needed to prevent epidemics. Application of biological or chemical substances has been seen as essential to reduce catastrophic diseases. However, most of these methods seem to have no clear benefit to this industry.8 To some extent progress has been made. These include but are not restricted to widespread use and standardization of diagnostic tests, wider application and improvement of biosecurity, and more effort on epidemiology and molecular studies of microbial dynamics in ponds and tanks.58–59 However, some serious problems still remain to be resolved since infectious pathogens continue to emerge in aquaculture facilities including ponds, tanks and hatcheries.
Currently, the immune system of these aquacultured animals has become the prime interest for many researchers to overcome the problem. Knowledge on host–pathogen interactions at the molecular level may provide a better and more effective management strategy to combat the catastrophic impact of infectious pathogens in order to maintain or increasing prawn production. Therefore, the high throughput molecular method, suppression subtractive hybridisation seems essential in elucidating pathogen genomes and the differentially expressed genes in particular immune–related genes of the host during infection.
Suppression subtractive hybridization is a powerful method that offers many advantages in disclosing differentially expressed genes in any given sample. Despite its advantages, this technique also has a drawback that genes with limited poly(A) or with mRNA which is not quantitatively altered by the vent in question cannot be detected. Therefore, several methods have been combined with SSH to circumvent this limitation. In respect to gene studies in prawns, this technique has opened a great opportunity to identify functionally important genes of penaeid species. Many genes have been identified from SSH cDNA libraries of penaeid prawns affected by viral, bacterial and environmental challenges. However, many aspects of immune–related genes in penaeid prawns including the regulatory mechanism, the exact role, and the effect of alteration of these genes in immunity of the penaeid prawns are still awaiting further investigation.
None.
None.

©2018 Rusaini,. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.