Journal of
eISSN: 2378-3184


Research Article Volume 2 Issue 1
1Department of Animal Husbandry and Fishery, Tadulako University, Indonesia
1IO-BAS Bulgarian Academy of Sciences, Bulgaria
2College of Public Health, Medical and Veterinary Sciences, James Cook University, Australia
2LAMANS Management Services S.A, Greece
Correspondence: Leigh Owens, College of Public Health, Medical and Veterinary Sciences, James Cook University, Townsville, Queensland 4811, Australia, Tel 61 7 4781 4632
Received: December 15, 2014 | Published: March 14, 2015
Citation: Rusaini, Owens L (2015) Expressed Genes in the Lymphoid Organ of Broodstock Banana Prawn (Penaeus merguiensis) Using Suppression Subtractive Hybridization. J Aquac Mar Biol 2(1): 00016 DOI: 10.15406/jamb.2015.02.00016
Most systemic viral infections in penaeid prawns are associated with the formation of lymphoid organ spheroid (LOS) cells in the lymphoid organ (LO). The development of spheroids is suggested to be a major defence mechanism against viral infection in penaeids. Spheroids were observed in a broodstock population of banana prawns (Penaeus merguiensis), while these changes were not identified in the wild population. This opportunity was taken to see how well suppression subtractive hybridization (SSH) would resolve a histological change with no known aetiology. A total of 316 sequenced clones were clustered into 141 contigs and 51.6% of the clones shared significant similarities to known peptide or nucleotide sequences in the NCBI GenBank database. These transcripts were assigned into 8 categories, including immune related genes (2.5%), synthesis, processing and regulation-related proteins (4.4%), ribosomal proteins (6.3%), proteases and inhibitors (7.0%), energy and metabolism factors (7.3%), structural and cytoskeletal related proteins (10.1%) and other sequences (13.9%). Many sequenced clones (48.4%) from the libraries had no significant similarity to amino acids/nucleotides in the public database revealing the ability of this method in disclosing new differentially expressed genes in the lymphoid organ of prawns. Despite the detection of differential gene expression in the SHH libraries, unfortunately there were no viral genomes identified which were associated with spheroid formation within the LO of penaeids. However, polymerase chain reaction (PCR) amplification with HPV140F and HPV140R primers suggested that the development of spheroid cells in the lymphoid organ of the broodstock population most likely due to Penaeus merguiensis hepandensovirus (PmeDV) which maybe the senior synonym of lymphoidal parvovirus (LPV).
Keywords: Banana Prawn, Penaeus merguiensis hepandensovirus, Lymphoid Organ, Spheroid Cells, Suppression Subtractive Hybridisation, Expressed Genes
LOS, Lymphoid Organ Spheroid; LO, Lymphoid Organ; SSH, Suppression Subtractive Hybridization; NCBI, National Centre For Biotechnology Information; PCR, Polymerase Chain Reaction; RT-PCR, Reverse Transcript-PCR; HPV, Hepatopancreatic Parvovirus; Pmedv, Penaeus Merguiensis Hapandensovirus; LPV, Lymphoidal Parvovirus; H & E, Haematoxylin and Eosin; RNA, Ribonucleic Acid; DNA, Deoxyribonucleic Acid; cDNA, Complementary DNA; BLAST, Basic Local Alignment Search Tool
The rapid development of the cultured prawn industry is associated with environmental and sociological disturbances in land use, the ecology of the aquatic organisms and global trade patterns. A major consequence of these changes is the emergence and the spread of infectious diseases. In the late 1980s, previously unknown diseases emerged in the cultured prawns both in Asia and Americas, spread rapidly to all countries farming prawns and brought catastrophe to this industry around the world. Even though some progress has been made to deal with these challenges and recovery has taken place in recent years, infectious diseases, in particular viral diseases still remain a major problem to the prawn aquaculture industry.1,2
Several viruses have been reported to cause diseases in the wild and cultured banana prawn Penaeus merguiensis including hepatopancreatic parvo-like virus (HPV) 3,4,5,6 the Australian strain of HPV (Penaeus merguiensis hepandensovirus, PmeDV) .7,8 lymphoidalparvo like-virus (LPV). 9 white spot syndrome virus (WSSV).10 and spawner-isolated mortality virus (SMV).11 Moreover, even though there is no report on banana prawns naturally infected with gill-associated virus (GAV), experimental infection suggested that this species may be susceptible to GAV 12
Histopathologically, most of these systemic viruses cause spheroid development within the lymphoid organ (LO) of banana prawns. The formation of spheroid cells in the lymphoid organ is a major defence mechanism to viral infection in penaeid prawns .13,14 Currently, in northern Queensland approximately 75-100% of cultured banana prawns from different family lines have lymphoid organ spheroid (LOS) cells that occupied more than 40% of the lymphoid organ area (Owens, unpublished data). However, no candidate virus has been identified in causing these spheroid formations.
Several studies have been conducted into disclosing differential gene transcripts of banana prawns. Most of published work on gene transcripts of P. merguiensis pays particular attention to the haemocytes. 15,16,17 and ovaries.8,19,20,21 Only a few studies on the tissue distribution of the genes including muscle, gonad, gills, brain, heart, intestine, hepatopancreas and lymphoid organ have been carried out.15,20 Furthermore, application of suppression subtractive hybridization (SSH) in banana prawns has only been performed to identify genes related to the ovarian development .18,20,21 Therefore, this study was conducted to determine the expressed genes in the lymphoid organ and detect the viral aetiology of spheroid cells in P. merguiensis using suppression subtractive hybridization (SSH).
Experimental Animals
Wild banana prawns Penaeus merguiensis that did not have spheroids in the lymphoid organ were caught with a cast net in creeks around Townsville, Queensland, Australia. Broodstock P. merguiensis that did have spheroids in the lymphoid organ were sourced from a hatchery in northern Queensland where the lymphoid organs were extracted in situ. Prior to histological examination and LO extraction, prawns were anesthetised by placing them in iced water for few minutes.
Histology
Experimental prawns were fixed in Davidson’s fixative for 48 hours. The cephalothorax was cut mid sagitally, placed in a histocassette and preserved in 70% ethanol and then processed for routine histological examination using standard paraffin embedded protocol. 22 Paraffin blocks were cut at 5 μm and stained with haematoxylin and eosin (H & E) and examined under a light microscopy.
Suppression Subtractive Hybridization
Twenty Broodstock P. merguiensis with an average body weight of 38.8 ± 6.8 g (SD) from a broodstock population and 20 banana prawns with a mean body weight of 12.0 ± 5.2 g from the wild caught population were used for the suppression subtractive hybridization. Total RNA was separately extracted from the lymphoid organ of wild and hatchery populations using SV Total RNA Isolation System (Promega) following the protocol of the manufacturer. Polyadenylated (poly(A)+) RNA was isolated from pooled total RNA using PolyATract® mRNA Isolation System III (Promega) as described by the company. Prior to use, 1000 μl poly(A) RNA was concentrated by freeze drying (Telstar 23750 - Cryodos -50/230 V 50 Hz, the UK), re-dissolved in25 μl RNAase-free water and kept at -80°C until used. The concentration and purity of nucleic acids were determined using spectrophotometry (NanoPhotometerTM, Implen, Germany).
Suppression subtractive hybridization was performed using the PCR-Select cDNA Subtraction Kit (Clontech, USA) following the manufacturer’s instructions to generate cDNA forward libraries between Broodstock prawns (tester) and wild caught prawns (driver). The tester cDNA was prepared from 1.19 μg of poly (A)+ RNA and the driver cDNA was synthesised from 0.02 μg of poly (A)+ RNA. Tester and driver cDNAs were digested with Rsa I to obtain shorter blunt-ended molecules. Rsa I-digested tester cDNA was divided into two portions. One portion was ligated with adaptor 1 and another one with adaptor 2R. In the first step of hybridization, an excess of driver cDNA was mixed with each tester cDNA, heat denatured and allowed to anneal to generate several fractions of molecules. In the second hybridization step, the two tester cDNAs from the first step were mixed together in the presence of freshly denatured driver cDNA to further enrich the differentially expressed genes. Finally, two rounds of PCR were performed to exponentially amplify the differential gene expression and suppress the common genes of the two cDNA samples. The subtracted PCR products were then inserted into pGEM-T easy vectors (Promega) and transformed into JM109 competent Escherichia coli cells which were plated onto agar containing ampicillin, X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) and IPTG (isopropyl-β-D-thiogalactopyranoside). Plasmid DNA was isolated from randomly selected white colonies and commercially sequenced by Macrogen Inc., Korea. Nucleotide sequences were analysed with BLASTx and BLASTn against known amino acid/nucleotide sequences on public databases (NCBI). Sequences with E-values <1e-05 were considered significant.
Reverse Transcriptase - Polymerase Chain Reaction (RT-PCR) Amplification with Bunyavirus Primers and PCR Amplification with Parvovirus Primers
To investigate the possibility of the aetiological agent of spheroid cells in P. mergueinsis was caused by virus with no poly (A) tail, several primers sourced from related-genus Phlebo virus within Bunyaviridae family including Mourilyan virus (AY927991), Uukuniemi virus (M17417) and Toscana virus (EU003175) were designed to amplify possible viral genomes in the Broodstock population. Total RNA was extracted from the lymphoid organ of Broodstock P. merguiensis (20 prawns) using SV Total RNA Isolation System (Promega). Complementary DNA (cDNA) was synthesised from total RNA using random hexamer primers (SuperScriptTM III first-strand synthesis system for RT-PCR, Invitrogen, USA). The RT-nested PCR was performed using primers MoV24F and MoV25R in the primary PCR, and primers MoV148F and MoV149R in the nested PCR (Table 1).
|
Primer name |
Sequences (5' to 3') |
Expected amplicon (bp) |
GeneBank ID |
References |
|
Bunyavirus Primers |
||||
|
MoV24F |
GGG ATG GTG TTG CCA TAC AAA GG |
610 |
AY927991 |
Cowley et al.23 |
|
MoV25R |
GTC ATT AGC TGG TCT TAG TTT TCA C |
|
|
|
|
MoV148F |
ACA GTT TGT CAA GCT CAC AGG ATG |
322 |
AY927991 |
Cowley et al.23 |
|
MoV149R |
AGA AGC GCC ATT CTG ATG AAC ATC |
|
|
|
|
MoV210F |
GGC CAC CCT TAC TAT CCT TG |
249 |
AY927991 |
Present study |
|
MoV439R |
ATT GTC CTT GTC TCG GGG TC |
|
|
|
|
UUKV2558F |
TTC CAA TAA GTG TAG CCC AAG |
668 |
M17417 |
Present study |
|
UUKV3205R |
AAA GAC ACG GCT ACA TGG AAC |
|
|
|
|
TosV2667F |
AGC GAA AAG CAA TTT ATC TCA |
416 |
EU003175 |
Present study |
|
TosV3064R |
CTC ATA GCC ATC AGA ACC A |
|
|
|
|
Parvovirus Primers |
||||
|
HPV140F |
CTA CTC CAA TGG AAA CTT CTG AGC |
140 |
DQ458781 |
La Fauce et al.7 |
|
HPV140R |
GTG GCG TTG GAA GGC ACT TC |
|
|
|
Table 1 Primer sequences used to amplify presumptive Bunyavirus and Parvovirus in the lymphoid organ of a Broodstock population of Penaeus merguiensis
In the primary amplification, PCR mixture contained 12.5 µl of GoTaq® Green Master Mix (Promega, USA), 1 µl of DNA template, and 0.75 µl (10 µM) of each primer. This PCR reaction volume was adjusted with nuclease free water (Promega) to a final volume of 25 μl. The PCR amplification was performed in a Master cycler gradient 5333 (Eppendorf, Germany) with an initial denaturation 95°C for 2 mins, 35 cycles for 30s denaturation at 95°C, 30s annealing at 60ºC, 40s extension at 72°C, and then followed by final extension for 7 mins at 72°C. In the nested PCR amplification, 2 μl of primary PCR products was amplified using primers MoV148F and MoV149R in 25 μl reaction mixture. Amplification profile was different from the primary PCR with annealing temperature at 58ºC and a shorter extension time for 30s.23 For other primers including primers TosV2667F/TosV3064R, the reaction mixture was the same as described for the primary PCR of primers MoV24F/MoV25R but it was slightly different in amplification profile. Amplification profile of primers MoV210F/MoV439R and primers UUKV2558F/UUKV3205R consisted of initial denaturation at 94°C for 7 mins, 40 cycles for 45s denaturation at 94°C, 45s annealing at 58ºC, 1 min extension at 72°C, and then followed by final extension for 5 mins at 72°C.
Deoxyribonucleic acid (DNA) was isolated from the LO of Broodstock banana prawns (20) using a High Pure PCR Template Preparation Kit (Roche Diagnostics), while DNA from the LO of wild population (20 banana prawns) was extracted using Wizard® SV Genomic DNA Purification System (Promega). Using HPV140F/HPV140R primers, the PCR product was amplified at 94°C for 7 mins for initial denaturation, 40 cycles for denaturation at 94°C for 45s, annealing at 65ºC for 45s, extension at 72°C for 1 min, and then finally subjected to extension at 72°C for 5 mins .7 Amplified products (10 µl) were visualised on a 1.2% agarose-TAE gels containing GelRed (10,000× in water) at a concentration of 0.5:10,000. Gels were visualised and photographed using InGenius LHR, gel documentation and analysis system (Syngene, UK).
Histology
Routine histological examination with H & E stain (Figure 1) showed that 12 out of 12 of Broodstock P. Merguiensis had lymphoid organ spheroid cells, while these pathological changes were not observed in the lymphoid organ of wild caught prawns (12/12). These abnormal cells had more basophilic cytoplasm and had no central lumen compared to the normal stromal matrix cells of the lymphoid organ. Some spheroids were bounded by elongated flattened cells or fibrous connective tissue and had cytoplasmic vacuoles.
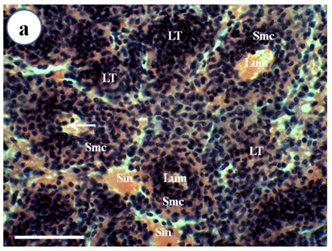
Figure 1a Normal lymphoid tubule (LT) without lymphoid organ spheroid (LOS) cells of wild caught P. merguiensis. Lymphoid tubules consist of lumen (Lum) surrounded by stromal matrix cells (Smc). Haemocytes (arrow) sometimes can be observed within the tubular lumen.
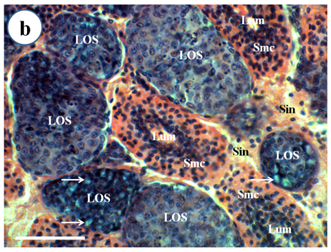
Figure 1b Spheroid development within the LO of broodstock P. mergueinsis. Spheroid cells are formed in the haemal sinuses (Sin) and appear to have a more basophilic cytoplasm and lack of a central lumen compared to the normal lymphoid tubule. Some spheroids demonstrated cytoplasmic vacuoles (arrow). H & E stain. Scale bar: 50 µm.
Figure 1 Light micrograph of longitudinal section of the LO of Penaeus merguiensis.
Suppression Subtractive Hybridization
Two bands with approximate 250 bp and 200 bp were visualised following gel electrophoresis and these PCR product was used as SSH libraries. Following hand picking of selected white colonies, 328 clones were sequenced and a total of 316 sequences were clustered into 141 contigs (consensus sequences) with a range of fragment sizes between 47 bp and 427 bp. Homology search revealed that around 51.6% of the total clones (163 out of 316 clones) shared significant similarities to known amino acids or nucleotides in the NCBI GenBank database. Transcripts were assigned functions as predicted from sequence homology from the public database and grouped into 8 categories (Figure 2). Many sequenced clones (48.4%) from the SSH libraries had no significant similarity to amino acids/nucleotides in the public database. This indicates the ability of SSH method in revealing new differentially expressed genes in the lymphoid organ of penaeid species.
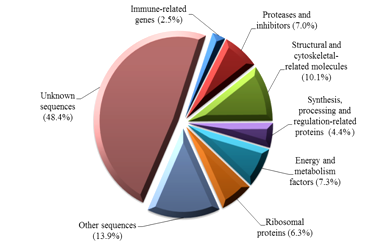
Figure 2 Functional categories of differentially expressed genes from the lymphoid organ cDNA suppression subtractive hybridisation (SSH) libraries of banana prawn, Penaeus merguiensis.
Reverse Transcriptase - Polymerase Chain Reaction (RT-PCR) Amplification with Bunyavirus Primers and PCR Amplification with Parvovirus Primers
Primary PCR with MoV24F/MoV25R primers produced an approximate 300 bp amplicon from 1 out of 20 samples examined. In the nested PCR, this sample also had an approximate 300 bp amplicon size. However, sequencing results failed to show any similarity to Mourilyan virus or other viral sequences, but similarity with zebra fish DNA (BX248086). Primers MoV210F/MoV439R generated expected amplicons from 2 samples and other different amplicon sizes from several samples. However, once again sequencing results showed no similarity to viral sequences. Primers UUKV2558F/UUKV3205R and TosV2667F/TosV3064R produced no amplicons.
Interestingly, when HPV140F/HPV140R primers were applied, three bands with approximate amplicon sizes of 140 bp, 200 bp and 250 bp appeared in the electrophoretic gel (Figure 3) from DNA templates of the Broodstock population. From the DNA templates of wild caught prawns, only two bands with approximate ampilicons sizes of 200 bp (4 samples) and 250 bp (5 samples) were observed in the electrophoretic gel. Since the 140 bp was the expected amplicon size for these primers, then three of these four bands (from DNA templates of prawns from the Broodstock population) were extracted from the gel, purified, cloned and sequenced. The sequencing results revealed that the transcripts had 100% (5e-42) nucleotide similarity to Australian Penaeus merguiensis densovirus, PmergDNV (DQ458718) that is currently called Penaeus merguiensis hepandensovirus (PmeDV).24

Figure 3a
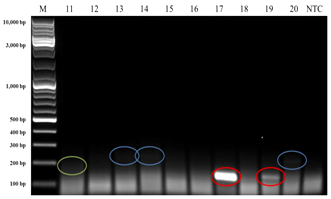
Figure 3b
Figure 3 PCR using HPV140F and HPV140R primers produced approximate 140 bp (red highlight), 200 bp (green highlight) and 250 bp (blue highlight) amplicon sizes from broodstock (hatchery maintained)Penaeus merguiensis. Lane M = 10 kb DNA ladder (GeneRulerTM, Fermentas).
NTC: Non-Template Control
Suppression Subtractive Hybridization
Sequenced clones from the lymphoid organ cDNA SSH libraries were clustered into several functional groups. These genes represented immune-related genes, synthesis, processing and regulation-related proteins, proteases and inhibitors, structural and cytoskeletal related proteins, energy and metabolism factors, and ribosomal proteins, which are all known to be involved in biological process and immune response against infectious pathogens and mostly play an important part in antiviral defence mechanisms (Table 2).
|
Genes |
Putative functional activities |
||
|
Viruses |
Bacteria |
Biological process |
|
|
Heat shock protein (HSP) |
Rungrassameeet al.93 |
||
|
Gamma-interferon-inducible lysosomal thiol reductase (GILT) |
Kongtonet al.73 |
Kongtonet al.72 |
Hastings and Cresswell 65 |
|
Anti-lipopolysaccharide factor (ALF) |
― |
||
|
Cathepsin |
― |
― |
Mort and Buttle.80; Aokiet al.52; Wanget al. 102; Stephenset al.96; Turket al.97 |
|
Metallothionein (MT) |
Ilbacket al. 69 |
― |
|
|
Calreticulin precursor (CRT) |
Chenet al[57 |
||
|
Eukaryotic translation elongation factor (eEF) |
Kawaguchiet al.71 |
― |
|
|
Eukaryotic translation initiation factor (eIF) |
Wanget al.,101 |
― |
|
|
Protein disulfide isomerase (PDI) |
Wilkinson and Gilbert, 104 |
||
|
Ubiquitin (Ubq) |
― |
Pickart and Eddins, 85 |
|
|
Signal peptidase complex (SPC) |
Paetzelet al. 82 |
― |
|
|
Cytochrome c oxidase (CO) |
Leuet al.74; Prapavoraratet al.,88; Liuet al.76; Pongsomboonet al. 86 |
Capaldiet al. 55 |
|
|
Mitochondrion |
― |
||
|
NADH dehydrogenase |
Heet al. 66 |
||
|
Actin |
Chongsatjaet al. 59; Leuet al. 74; Wuet al.106; Pongsomboonet al. 86 |
Zhanget al.107 |
Hildet al.68 |
Table 2 Genes that may putatively be involved in a range of functional activities identified in cDNA suppression subtractive hybridization (SSH) libraries of the lymphoid organ (LO) of banana prawn, Penaeus merguiensis.
Putative functions against pathogens are assigned based on the up regulation of the genes reported in the published work as indicated. Dash (-) indicates no data available in the published literature.
Sequence with homology to viral genes was not identified in the SSH libraries. There are several possible reasons why sequences with homology to viral genes was absent in the SHH libraries. Firstly, the abundance of viral transcripts in the tester cDNAs (Broodstock population) was too low to be expressed using SSH. The abundance of the viral cDNA should be more than 5-fold concentrated in the tester than in the driver or the fractional concentration of mRNA of the viral gene needs to be at least 0.1% of the total mRNA in order to be detected using this method, because the completion of the second hybridization cannot be achieved if the target cDNA is too low 25,26. Secondly, the poly (A) tail on the mRNA of the virus may be too short for this SSH technology to be successful. Genes in target should have at least four or more poly(A)s in the tail in order to be amplified with SSH technique, because the complementary DNA synthetic primer of this protocol contains four poly(T)s at the first 5΄ end 27. Finally, the virus causing these cellular changes may not have a poly(A) tail, therefore it could not be expressed in the SSH libraries 26. It should be noted that ~40% of the genes of WSSV do not have a poly(A) tail but were expressed 28. This is why the RT-PCRs for bunyavirus were attempted and further studies are necessary to determine which hypothesis is more likely (see below).
Reverse Transcriptase - Polymerase Chain Reaction (RT-PCR) Amplification with Bunyavirus Primers and PCR Amplification with Parvovirus Primers
The possibility of the viral aetiology of the spheroids in P. merguiensis due to virus with no poly(A) tail, was investigated using primers designed from related-genus Phlebo virus of family Bunyaviridae. Bunyaviral genome comprises negative-sense tripartite single-stranded RNA (ssRNA) and the viral mRNAs do not have a poly (A) tail 29,30. In addition, wild and farmed P. monodon and P. japonicus were naturally infected with Mourilyan virus (MoV) 31 and a quite broad host range may have been infected with this virus (Cowley, pers. commun.). However, the lack of positive results showed that these lymphoid organ changes were unlikely to be associated with bunyaviruses.
Hepatopancreatic parvovirus 140 (HPV140) primers were then used to amplify the suspected viral genomes which produced interesting results. This suggested that the formation of spheroid cells in the lymphoid organ of the hatchery prawns was related to PmeDV infection and these cellular changes may be formed as defensive response against this viral infection. The absence of viral (PmeDV) transcripts from cDNA SSH libraries is most likely due to the low viral load in the LO of the tester population. The presence of only four positive samples on the PCR amplification suggested that in chronically infected prawns, either PmeDV was undetected due to low viral load in the remaining samples or this virus has been eliminated during spheroid cell development by an unknown mechanism. 32 Hasson et al. 33 who used in situ hybridization in the chronic phase of viral infection believed that rapid development of massive spheroid cells within the LO only contained low grade to moderate number of virions in infected foci. According to Tang and Lightner 34, HPV is an enteric virus targeting hepatopancreatic and intestinal cells of penaeid prawns. Thus, PmeDV could be an enteric virus that spills over into the open circulatory system of all systematic tissues including the LO of the prawns. Indeed, systemic circulating PmeDV has been found in the mud crab, Scylla serrata .35 In addition, another reported parvovirus of penaeid prawns, lymphoidalparvo-like virus (LPV) that was found in P. merguiensis, P. monodon and P. esculentus also infected systemic tissues including antennal gland, nerve cord and lymphoid organ. Based on the molecular evidence presented here and the principle of Ockham’s razor (the law of parsimony), it is highly probable that LPV recorded by Owens et al. 9 was indeed systemic PmeDV.
Furthermore, the formation of spheroids within the lymphoid organ has been detected in many naturally or experimentally infected penaeid species with viral diseases (Rusaini and Owens, 2010). These viruses included LPV .9 lymphoid organ vacuolization virus (LOVV).36, rhabdovirus of penaeid shrimp (RPS).37 lymphoid organ virus (LOV)/GAV .38,39 and SMV .40 The spheroid cell formation has also been implicated to be associated with YHV.41,42 TSV 43, WSSV 44,45,46 infectious myonecrosis virus (IMNV).47 Mourilyan virus (MoV) .23,48 and Laem-Singh virus (LSNV).49
To conclude, the massive development of spheroid cells in the lymphoid organ of P. merguiensis from the broodstock population seems most likely to be due to defence mechanism to viral (PmeDV) infection. As a result, the health status of these two populations of banana prawn was remarkably different causing differential gene expression between populations, with some genes being induced in the Broodstock population. The up-regulation of these genes implicated their involvement in the immune responses. All in all, the current investigation has provided some valuable evidence on the up regulated genes in the lymphoid organ that may play crucial roles in viral defence responses in penaeid prawns. This could be used for further research on the host-viral interaction leading to a new immune-intervention approach that may help to circumvent the catastrophe of viral diseases in penaeid prawn industry.
The authors would like to thank Dr. Kathy La Fauce for reading the manuscript and offering valuable suggestions on the bench work. We also acknowledge northern Queensland prawn farmers for their excellent cooperation in supplying Penaeus merguiensis Broodstock for this research. This work partly supported by Graduate Research Scheme (GRS) Grant 2010 Faculty of Medicine, Health and Biomolecular Sciences, James Cook University. Rusaini was a recipient of Australian Development Scholarship (ADS) Program from Australian Agency for International Development (AusAID).
None.

©2015 Rusaini,. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.