Journal of
eISSN: 2572-8466


Research Article Volume 10 Issue 6
Instituto de Investigaciones Químico-Biológicas de la Universidad Michoacán de San Nicolás de Hidalgo, México
Correspondence: Beltrán-Peña E., Laboratorio de Transducción de Señales, Edificio B3, Instituto de Investigaciones Químico-Biológicas de la Universidad Michoacán de San Nicolás de Hidalgo, Ciudad Universitaria, Morelia, 58030, México
Received: November 10, 2023 | Published: November 24, 2023
Citation: Carrillo-Flores E, Mellado-Rojas ME, Beltrán-Peña E. Plasmodesmata role on plant development regulation. J Appl Biotechnol Bioeng. 2023;10(6):171-176. DOI: 10.15406/jabb.2023.10.00345
Plants are sessile organisms that depend on the root system that anchors them to the soil and it permited to taken water and nutrients. Root system development depends on natural auxin, indole-3-acetic acid. The auxin are transported in plants by the polar auxin transport (PAT) and the symplastic transport (ST) through of the plasmodesmata (PD). In the present work, the participation of the TS during the development of A. thaliana was analyzed.
Keywords: callose, plant development, plasmodesmata, polar auxin transport, symplastic transport
ABCB/PGP, ATP-Binding Cassette/P-GlycoProtein; AUX/IAA, Auxin/Indole-3-Acetic Acid; ARF, Auxin Response Factor; CALS/GSL, callose synthase/glucan synthase-like; GFP, Green Fluorescent Protein; GH3, GRETCHEN HAGEN3; IAA, Indol-3-Acetic Acid; IPA, Indole-3-Pyruvic Acid; LR, Lateral Root; LRP, Lateral Root Primordia; PAT, Polar Auxin Transport; PIN, PIN-FORMED efflux; PDLP, PlasmoDesmatal-Located Protein; PD, PlasmoDesmata; PDCB, PD-Callose Binding protein; PR, Primary Root; QC, Quiescent Center; RAM, Root Apical Meristem; SAUR, Small Auxin-Upregulated RNA; SEL, Size Exclusion Limit; SCN, Stem Cell Niche; SKP1, S-phase Kinase-associated Protein1; CUL1, CULLIN1; RBX1, ring box protein1; ST, Symplastic Transport; TAA, Trp Aminotransferase; TIR1, Transport Inhibitor Response1; Trp, Tryptophan
The natural auxin, indole-3-acetic acid (IAA), which regulates practically all plant development processes, is synthesized in plant foliage and mobilized to sink tissues by two types of transport; a rapid through of phloem and another that is carried out cell-cell and it known as polar auxin transport necessary for the auxin gradients establishment.1 Recently, it has been shown that auxin is also mobilized through channels calls plasmodesmata, movement known as symplastic transport that complements at PAT.2 Studies several indicate that plants close PD due to callose deposition that causes a restriction of ST of auxins, affecting stomata development, lateral roots and plants tropic responses, which this manuscript will review the ST importance in plant development.
Auxin response network
The word auxin comes from the Greek auxein which means "to grow" and was discovered by observing of coleoptile bending of Phalaris canariensis towards light. Years later, it was shown that this effect was due to auxin accumulation in area of tissue folding.3 It is now known that virtually all plant development is auxin regulated.4–8 The auxin response network comprises the following events: i) homeostasis, ii) transport and iii) perception and signaling9 which are described below. i) Homeostasis, maintains the internal balance of IAA, through multiple and dynamic adjustments in synthesis, storage, catabolism and conjugation of IAA. In Arabidopsis, IAA biosynthesis from tryptophan (Trp) can be carried out by four different pathways.10–13 The pathway main involved at indole-3-pyruvic acid (IPA), where Trp is converted by trp aminotransferases (TAA) to IPA, which at the same time is transformed by flavin mono-oxygenases YUCCA (YUC) to IAA (Figure 1).14 ii) IAA is distributed in plant by two ways: one from the young leaf tissues where it is synthesized to sink tissues through the phloem and another through of PAT over short distances. The PAT requires influx carriers AUXIN1/LIKE-AUX1 (AUX1/LAX) and PIN-FORMED efflux (PIN) and ATP-Binding Cassette/P-glycoprotein (ABCB/PGP) (Figure 1).15 iii) In perception and signaling events, when auxin concentration is low, the response is inhibited by the repressors Auxin/Indole-3-Acetic Acid (AUX/IAA) that sequester at the Auxin Response Factor transcription factors (ARF) and thereby prevent the expression of auxin early response genes. While, at auxin high concentrations it is perceived by nuclear receptor Transport Inhibitor Response1 (TIR1) binding to SCF complex (S-PHASE KINASE-ASSOCIATED PROTEIN1 (SKP1), CULLIN1 (CUL1), RING BOX PROTEIN1 (RBX1) and a F-box ligase E3 of ubiquitin, this allows auxin to bind AUX/IAA to the complex SCF. When this happens, the AUX/IAA are marked by the E3 ubiquitin ligase and it degraded via proteasome, with the consequent release of the ARFs and genes expression: Small Auxin-Upregulated RNA (SAUR), GRETCHEN HAGEN3 (GH3) and AUX/IAA (Figure 1).16–18 As articles numerous have shown PAT participation during the plant development 19,20 and recently it has been reported that auxins, in addition to moving through the PAT, also diffuse through ST21–24 whereby both processes are described below.
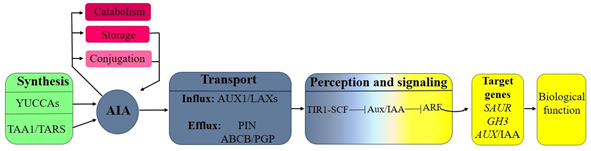
Figure 1 Auxin response network: The balance between auxin synthesis (green box), conjugation, storage and catabolism (rose boxes) controls the pool of IAA (blue disc). Cellular auxin content also depends on the auxin transporters (blue box). Binding of auxin to the SCFTIR1/AFB auxin receptor complexes, allows ubiquitination and degradation of the Aux/IAA protein, depressing activating ARF-regulated loci (yellow boxes).24
Polar auxin transport
In Arabidopsis thaliana the family of PIN efflux carriers consists of members eight. At primary root (PR) tip, auxins that arrive through the phloem are redistributed by PIN carriers different, creating whit it an auxin maximum in the Root Apical Meristem (RAM), essential for the indeterminate growth of root. In Figure 2A we can observed that PIN1 participates in auxin movement through vascular bundle; PIN4 it concentrates in Quiescent Center (QC); PIN3 and PIN7 redistribute them in region of columella and PIN2 transports the auxins from lateral root cap to epidermis and the same carrier returns it through cortex towards the QC, maintaining an auxin maximum at root tip, necessary to supported the Stem Cell Niche (SCN) function. SCN is formed by QC surrounded by initial cells it which will give rise to cell types different that form the root (Figure 2B). It has been reported that efflux carriers PIN1 and PIN3 are involved in lateral roots (LR) development (Figure 2A).25,26
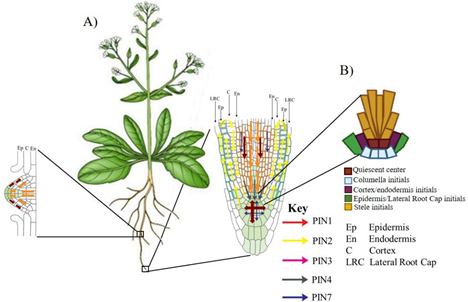
Figure 2 Auxin transport by the PINs carriers in Arabidopsis root: (A) At primary root tip the auxins are mobilized by PINs different, while only two members of PIN participates in root lateral development. (B) Schematic of Stem Cell Niche (SCN) of the primary root of Arabidopsis.27
Auxin symplastic transport through of the plasmodesmata
We can has observed in Figure 3A that auxins in addition to being mobilized through PAT, also it diffuses through plasmodesmata by the symplastic transport. It has been reported that the ST of auxin affects phototropism, lateral root emergence, and leaf hyponasty.23 Mellor, et al.,2 compared in vivo experimental data of Arabidopsis root auxin level with the level predicted by in silico experiments (Figure 3B). The results from both distributions showed very large discrepancies between auxin concentrations when only PAT was considered, unlike the high agreement when both PAT and ST were taken into account. This allowed the authors to recommend that in any process of plant development that involves the auxin transport, both types of movement must be considered.
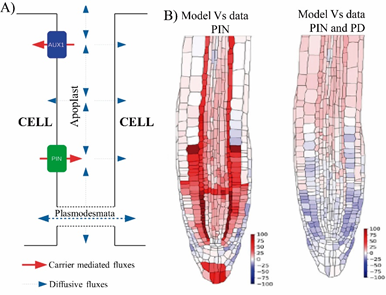
Figure 3 Components that modulate auxin movement through the polar transport and symplastic transport. (A) Schematic of the auxin fluxes. Although auxin is transported via influx and efflux carriers, the presence of PD enables diffusion of auxin directly in adjacent cells. (B) Differences between in silico distribution and experimental data with the DII-VENUS reporter line that allow us to observe auxin level in Arabidopsis root tip. Discrepancies between auxin concentrations, when only auxin transport by PINs is considered, are indicated in colors ranging from pink to red (left). While the coincidences, when both, the movement of auxin through the PINs and the diffusion through the PD are taken into account are represented in color from white to lilac (right).2
Plasmodesmata structure and regulation of its opening and closing
The PD are nanoscopic channels that pass through the cell wall and cytoplasm neighboring cells connect. The molecules movement through of PD depends on their permeability, characteristic known as size exclusion limit (SEL), which is defined by the maximum size of a molecule capable of passing through PD28–31 and that depends on concentration gradient between adjacent cells. Proteins, RNAs, viruses, IAA and molecules of up to 80 kDa are mobilized through these PD.32 The PD permeability is regulated by mechanisms dependent and independent of callose (polymer of glucose linked by β-1,3 bonds) deposited in PD necks (Figure 4). The dependent mechanisms involve synthesis and degradation of the callose, while that it independent included the alteration in the frequency and changes in PD structure from simple to branched.33 Callose synthesis is carried out by callose synthases, CALLOSE SYNTHASE/GLUCAN SYNTHASE-LIKE (CALS/GSL) and it has been observed that callose levels high on PD neck close it and ST restrict. Proteins that help callose deposition on PD neck also participate in PD closure, such as PD-CALLOSE BINDING PROTEIN (PDCB)34,35 and PLASMODESMATAL-LOCATED PROTEIN (PDLP) (Figure 4B).36,37 On the other hand, callose removal that results in PD opening is mediated by β-1,3-glucanases (Figure 4A).38
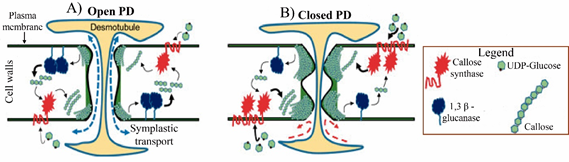
Figure 4 Callose mediated plants open and close plasmodesmata: (A) A cartoon representing ‘open’ plasmodesmata (left). (B) ‘Closed’ plasmodesmata (right) due to over accumulation of callose in cell walls.38
Callose participation on defense response plants
The plants defense response against invading pathogen was one of reported functions first of PD, because viruses, fungi and bacteria use PDs to invade them.39,40 To face the attack of pathogens, the plants expression increase of CALS/GSL, PDLP5, and PDCB and form a structure known as papillae that contains phytoalexins, reactive oxygen species, and defensins.41 Callose deposition in PD regulated by salicylic acid in response to pathogen attack, causing a decrease in SEL.42,43 An callose accumulation increase during the plant-bacteria interaction was reported in Arabidopsis exposed to Botrytis cinerea.44 In study another, was observed an callose increase in strawberry leaves exposed to the physical interaction and at volatile compounds of Botrytis methylotrophicus.45 The aforementioned results suggest that plasmodemal closure prevents that pathogen spread to plant rest.
Plasmodesmata role during plant development
Callose is synthesized by a multi-subunit complex, where the catalytic subunit callose synthase is the most important.46 The Arabidopsis genome contains twelve CALS/GSL callose synthase genes.47 The Arabidopsis gsl8 mutant prevents callose accumulation in PD and stomatal development impairs. As the gsl8 mutation caused other alterations on plant development, were created gsl8 mutant dexamethasone conditioned (dsGSL8 RNAi) to analyzed the effect of callose on hypocotyl response to light and gravity. It was observed that hypocotyls of dsGSL8 RNAi seedlings in dexamethasone presence did not present characteristic bending in response to both stimuli and showed an callose absence in bending zone. So results of these experiments indicated that callose accumulation was necessary for hypocotyl bending in response to the two tropisms evaluated.48
On the other hand, Vatén et al.,49 reported that CALS3 is expressed in vascular bundle and in RAM, and that roots of three gain-of-function mutants (it increased gene expression) in CALS3: cals3-1d, cals3 -2d and cals3-3d, collectively called cals3-d showed an aberrant pattern of movement marker, GFP encoded by pSUC2::GFP gene. In control seedlings pSUC2::GFP was mobilized in vascular bundle, while in homozygous line cals3-1d the marker disappeared and was partially restored in heterozygous line cals3-1d/+. cals3-d seedlings showed an extremely short primary root compared to wild type and when analyzing the callose accumulation during LR development with aniline blue, the mutant showed a greater callose accumulation respect to Wt. The lateral roots are induced from of structures known as lateral root primordia (LRP), whose development proceeds through of stages seven. In I stage, the founding cells of the pericycle are marked with auxins and later in the II-VII stages, cells it divide and form a dome that as it grows, successively breaks down the layers of the endodermis, cortex until reaching the epidermis. Interestingly, this development is regulated by auxins. Subsequently, auxins move towards tip the dome as it grows until they reach the meristems of the LR.50
As mentioned before, PDLP5 assists in callose deposition on PD neck and therefore contributes to plasmodesmal closure. Sager, et al.,51 observed that in I-II stages of the LRP development, PDLP5 are located in the endodermis, while in the IV-VI stages, in cortex and in epidermis when emerged the LR. Interestingly, they also observed that PDPL5 expression depends on auxin. The phenotype presented by the lines pdlp5-1 mutants and PDLP5OE overexpression was contrasting, pdlp5-1 showed a greater number of long LRs, while PDLP5OE seedlings presented a decrease in the PR and a growth of few LR and short. This behavior was attributed to an accelerated development of LRP in pdlp5-1 and a delay in PDLP5OE. Based on the aforementioned results, Sager, et al.,50 established a model that proposes that there is a transient symplastic isolation in some LRP development stages. In I-III stages, LRP is connected to phloem of the RP and PDLP5 is positioned on endodermis cells. In IV-VI stages, it is located in cortex, preventing GFP movement, while in VII stage, protein is positioned in epidermis, leaving at the LRP symplastically isolated from phloem of the RP. When the LR emerges, it develops its phloem and then symplastic connection with the phloem of RP is reestablished (Figure 5).
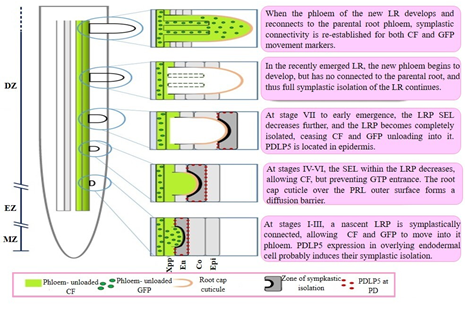
Figure 5 Progression of symplastic domains during lateral root primordia outgrowth.
Abbreviations: CF, CarboxyFluorescein; Co, Cortex; DZ, Differentiation Zone; En, Endodermis; Epi, Epidermis; EZ, Elongation Zone; LR, Lateral Root; LRP, Lateral Root Primordium; MZ, Meristematic Zone; PD, PlasmoDesmata; PDLP5, PlasmoDesmata-Located Protein 5; SEL, Size Exclusion Limit; Xpp, Xylem pole pericycle.52
Another example of callose participation in the LRs development is the behavior observed in the mutant double of glucanases, pdbg1,2, where the PDs are closed because there is not callose degradation. These seedlings presented a greater callose accumulation in vascular bundle of the PR, compared to Wt. Furthermore, these lines showed an increase in the lateral roots density. By observing these seedlings whit more detail, they noticed that at the site where normally forms one LRP, in these mutants appears several LRPs, resulting in a greater of lateral root number. The authors concluded that ST through PD is critical for the initiation and properly positioning of LRP.51,53
The root system is essential for plant anchoring to soil and for absorption of water and nutrients. So understanding molecular mechanisms that regulate root architecture is crucial to improve nutrient uptake efficiency and yield in crops of agronomic importance. The formation of the lateral roots is an essential organogenic process to establish the root architecture. Analyzes numerous have shown that auxins are central regulators of LR development. It has recently been reported that a transient symplastic isolation during the LRPs development and their subsequent symplastic reconnection to the PR phloem is an important event for the proper LRP development. In this review, several examples were presented that show that ST of auxin is a fundamental element not only for the establishment of root architecture but also for the development of other plant structures, as well as in the regulation of tropic responses of plants to light and gravity.
This work was supported by the Coordinación de la Investigación Científica of the UMSNH. E. C.-F. is fellow of CONAHCYT, México (801093).
The authors declare that there is not conflict of interest.

©2023 Carrillo-Flores, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.