eISSN: 2379-6367


Research Article Volume 8 Issue 3
1Department of Genetics, University of Bucharest, Romania
2Clinical laboratory–Bacteriology, Giurgiu County Emergency Hospital, Romania
Correspondence: Stanislav Alexandra Alina, Faculty of Biology, Department of Genetics, University of Bucharest, 1-3 Intr. Portocalelor, 060101, Bucharest 6th District, Romania
Received: April 23, 2020 | Published: June 1, 2020
Citation: Alina SA. The identification of the high risk of metabolic syndrome by applying MDR and Jalview bioinformatics programs. Pharm Pharmacol Int J. 2020;8(3):148-157. DOI: 10.15406/ppij.2020.08.00290
The metabolic syndrome (MS) is a complex multi-factorial disease associated with obesity (OB), type 1 diabetes mellitus (DM1), type 2 diabetes mellitus (DM2) and high blood pressure (HBP). This study included 404 subjects selected at Giurgiu Country Hospital, on the basis of clinical evaluation and of biochemical and hematological laboratory investigations, and the sequencing of 28 genes of interest: INS, IGF2, TGF-beta, HSPG Bam H1, ACE, UCP2, FABP2, PLIN1, PON1, FTO, ADRB3, BHMT2, MTHFR, IRS1, TRPM6, MT1A, APOA5, PEMT, ABCB4, CHDH, FADS2, PCYT1A, PCYT1B, PNPLA3, SCD, SCL44A1, STAT3 in identifying the high risk of developing metabolic syndrome. The findings were statistically processed by the MDR and Jalview programs.
Keywords: genes, MDR, Jalview, metabolic syndrome
The metabolic syndrome (MS) is a complex multi-factorial disease associated with obesity (OB), type 1 diabetes (DM1), type 2 diabetes (DM2) and high blood pressure (HBP). The metabolic and nutritional factors, complemented by the interactions between the genome and the environmental conditions mediated by epigenetic mechanisms, and play important role in the genesis of obesity, diabetes mellitus and high blood pressure.1 The purpose of study was to identify the risk of developing MS in patients diagnosed with obesity (OB), DM1, DM2 and high blood pressure (HBP) by using the MDR and Jalview programs, on the basis of clinical, laboratory (biochemical, hematological) and genetic data.
The 404 subjects, of whom 219 in patients with MS, OB, DM 2, DM 1, HBP and 185 clinically healthy subjects were enrolled in the study on the basis of their clinical evaluation: age, sex, height, weight, cardiac index, waist circumference, blood pressure, alcoholism, smoking, stress, diet, family history; biochemical laboratory investigations: glucose, triglycerides, HDL, cholesterol, LDL, HbA1c, creatinine, uric acid, urea, TGO, TGP, magnesium, calcium, folic acid, vitamin D, vitamin B12, homocysteine, vitamin C, which were determined by spectrophotometry; and hematological laboratory investigations: WBC, HGB, HCT, RBC. All the subjects were selected at Giurgiu County Emergency Hospital. An informed consent was obtained from all subjects before the commencement of the study. In isolating DNA from blood the DNeasy Blood and Tissue Kit from Qiagen was used;2 the spectrophotometric determination of the isolated genomic DNA purity and concentration3 and the sequencing of the risk genes were performed by Advanced NGx test.4 The statistical analysis was conducted through the following applications: Jalview Software,5 MDR Software.6 This study was conducted from 20.01.2014-19.01.2015 and 20.01.2015-19.01.2016 as part of a volunteer activity at Giurgiu County Emergency Hospital (Volunteer Contract no 2/20.01.2014 and contract no 2/20.01.2015 subject to art. 13(5) of Law no 677/2001).
The study included 404 subjects aged 21-92, of whom 219 inpatients diagnosed with MS, OB, DM 1, DM 2, HBP and 185 clinically healthy subjects from the hospital environment. It consisted in sequencing the genes of interest: UCP2, UCP3, FABP2, PLIN1, PON1, FTO, ADRB3, BHMT2, MTHFR, IRS1, TRPM6, MT1A, APOA5, PEMT, ABCB4, CHDH, FADS2, PCYT1A, PCYT1B, PNPLA3, SCD, SCL44A1, STAT3, INS, IGF2, ACE, TGF-beta, HSPG Bam H1 and studying their association with metabolisms in MS. By using the Advanced NGx test it was noted that the dominant homozygous and, respectively the heterozygous form for the adult nutrition included the markers of FADS2 gene associated with omega 6 and omega 3 unsaturated fatty acids (omega 6 and omega 3 are high concentrations );vitamin B2, vitamin B12, betaine, choline, magnesium and folates associated with the following genes: MTHFR, BHMT2, CHDH, PEMT, SLC44A1, TRPM6 (with high concentrations of homocysteine and low concentrations of folic acid, vitamin B12, magnesium, calcium, choline, betaine); genes PEMT, ABCB4, CHDH, FADS2, MTHFR, PCYT1A, PCYT1B, PNPLA3, SCD, SLC44A1, STAT3 are associated with non-alcoholic liver steatosis (with the occurrence of steatosis in overweight or obese individuals); the following genes: UCP2, UCP3, FABP2, PLIN1 were associated with obesity (the occurrence of obesity); genes BHMT2, MTHFR were associated with hyperhomocysteinemia (high concentrations of homocysteine and low concentrations of folic acid, vitamin B12, magnesium, calcium, choline, betaine, riboflavin (vitamin B2), curcumin, fish oil, vitamin C, vitamin D and vitamin E); cholesterol was associated with the following genes: UCP3, UCP2, PON1 (high concentrations of cholesterol, triglycerides, LDL-cholesterol and low concentrations of HDL-cholesterol); the following genes were associated with insulin-resistance: IRS1 and TRPM6 (high concentrations of glucose, HbA1c and magnesium); cardiovascular diseases were associated with gene MT1A (high concentrations of glucose and HbA1c); postprandial hyperlipemia was associated with gene APOA5 (high concentrations of cholesterol, triglycerides, LDL-cholesterol and low concentrations of HDL-cholesterol); and other genes associated with physical effort or performance, such as those for the cardiac and respiratory function, were associated with genes: SLC16A1, PPARA (high concentrations of cholesterol, triglycerides, LDL-cholesterol and low concentrations of HDL-cholesterol); the muscular function and the body weight were associated with genes: CHDH, FTO, ADRB3, ACE (deficiency of vitamin B12, folic acid, high concentrations of cholesterol, triglycerides, LDL-cholesterol, BMI, height, weight, glucose, HbA1c, systolic and diastolic blood pressure, and low levels of HDL-cholesterol); metabolism was associated with genes: UBE2E2, ADAMTS9-AS2, KLHDC5, IRS1, MEFV, MFE (high concentrations of glucose, cholesterol, triglycerides, LDL-cholesterol and low concentrations of HDL-cholesterol), which are associated with cardiovascular diseases, diabetes, obesity, and can increase the risk for metabolic syndrome.
It was noted that all these genes, with their genetic variations, associated with nutrition in healthy adults, and with lipid, carbohydrate and behavioral metabolism, were associated with a high risk of developing metabolic syndrome (Figures 1-13). After analysis of the secondary structure of the 28 genes of interest, it was noted that those genes, through the sequence of nucleotides preserved in position 1, were associated with the metabolic syndrome, by using Jalview 2.8 software.
The statistical analysis performed by applying the MDR software version 3.0.2. revealed that, in the whole set of clinical data, laboratory investigations and genes of interest investigated by Advanced NGx, OR was the highest, OR = 104.7455, OR > 1, which results in a high risk of developing metabolic syndrome (Figure 15). The Jalview 2.8 software was used to identify the secondary structure of genes UCP2, UCP3, FABP2, PLIN1, PON1, FTO, ADRB3, BHMT2, MTHFR, IRS1, TRPM6, MT1A, APOA5, PEMT, ABCB4, CHDH, FADS2, PCYT1A, PCYT1B, PNPLA3, SCD, SCL44A1, STAT3, INS, IGF2, ACE, TGF-beta, HSPG Bam H1. It was noted that the amino acid composition of the 28 genes was preserved and identical in position 1 of the nucleotide sequence. The phylogenetic tree was obtained by the Neiber Joining method. It was noted that genes PNPLA3, UCP3, HSPG, TRPM6 are phylogenetically related and they are composed of more sequences of common amino acids. FADS2, PON1, PLIN1, PCYT1A, PCYT1B, IRS1 are phylogenetically related and they are composed of fewer sequences of common amino acids. CHDH is phylogenetically related, but the amino acid sequences are different. APOA5, INS are phylogenetically close but the amino acid sequences are identical. UCP3, MTHFR, STAT3, TRPM6 are phylogenetically related, but they are different in sequences of common amino acids. MT1A, ADRB3, IRS1 are phylogenetically related and they are close in the common amino acid sequences. IGF2, ADRB3, TGFB, SCD, PNPLA3, FTO are phyolgenetically close and they are different in the common sequences of amino acids. ACE, INS, PEMT, SLC44A1, FABP2, UCP2, BHMT2 are phylogenetically related and they are similar in sequences of common amino acids. The genes were associated with the metabolic syndrome, obesity, diabetes mellitus and high blood pressure (Figures 14, 16).

Figure 12 Markers associated with the metabolism of vitamin B2, vitamin B12, betaine, choline, magnesium and folates.

Figure 14 A) Fasta format; B) Secondary structure; C) Composition of nucleic acids; D) Representation of the phylogenetic trees of genes UCP2, UCP3, FABP2, PLIN1, PON1, FTO, ADRB3, BHMT2, MTHFR, IRS1, TRPM6, MT1A, APOA5, PEMT, ABCB4, CHDH, FADS2, PCYT1A, PCYT1B, PNPLA3, SCD, SCL44A1, STAT3, INS, IGF2, HSPG, TGFb, ACE. PDB processed in Jalview program.
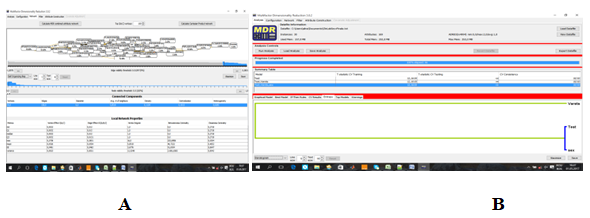
Figure 15 A) Graphic interaction model for MS, OB, DM1, DM2, HBP. B) Cluster by dendogram generated by MDR .
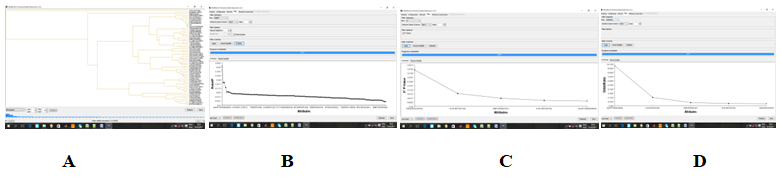
Figure 16 MDR data processing evaluated through (A) the dendogram provided by MDR c, (B) Relief F, (C) Val P, (D) OR
The 28 genes were associated with cardiovascular diseases, diabetes, obesity, which generates a high risk of metabolic syndrome. The Jalview 2.8 processing of the nucleotide sequences of the examined set of genes allowed the identification of secondary structure modifications in the corresponding proteins, which provides the premises for their association with possible functional alterations.The building of the phylogenetic tree by Jalview showed that the 28 genetic markers were associated with an increased risk of MS. The graphical model, the interaction map and the cluster obtained by applying MDR confirm that the polymorphisms of the 28 genes present an increased risk of MS. ReliefF=0,45, Value P=0,00001(p<0,0001), OR = 104.7455, OR > 1.
The application MDR was chosen to include all the investigated data: clinical, laboratory and genetic data for the identification of increased risk of MS.
Information about the investigated genes can be found in the published literature.
INS (insulin) gene. Cytogenetic location: 11p15.5.7 It plays an important role in producing insuline, which is necessary for the control of blood glucose.8 The genetic variant of insulin gene (VNTR and SNP) plays a role in the susceptibility to type 1 and type 2 diabetes mellitus. INS gene is associated with the metabolic syndrome,9 type 2 dibetes mellitus, polycystic ovary syndrome and coronary diseases.10
IGF2 gene (insulin-like growth factor 2). In humans, IGF2 gene is located on chromosome 11p15.5 in the region containing numerous imprinted genes.11 IGF2 is a regulator of somatic growth, cell proliferation and maternal imprinting (imprinting of the telomeric domain of region 11p15 containing silenced hypermetilated ASCL2, H19 and IGF2 in Wilms tumors).12 It is involved in stem cell differentiation.13 IGF2 influences placental lactation and can play a role in fetal development.14 The synthesis of several specific components such as proteoglycans and other components of the connective matrix. Using a feedback mechanism, it inhibits the release of the growth hormone (while stimulting the release of somatostatin).15 IGF2 acts as a growth factor and predisposes to diabetes mellitus. IGF2 gene is associated with obesity, type 2 diabetes mellitus16 and gestational diabetes.17
TGF-beta (Transforming growth factor beta) gene. Cytogenetic location : 19q13.1. TGF-beta fulfills important roles: it stimulates tissue repair and fibrosis by regulating cell proliferation and apoptosis and by synthetising components of the extracellular matrix; it participates in regulating the function of the immune system; it inhibits the functioning of T and B lymphocytes, the proliferation of neurons, of endothelial and mesenchymal cells, the synthesis of several cytokines;18 it plays a role in growth control, inflammation and tumorigenesis.19 TGF-beta gene was associated with type 2 and type 1 diabetes mellitus,20 obesity, diabetes mellitus, high blood pressure19 and MS.21
HSPG (Heparan sulfate proteoglycans) gene. Cytogenetic location : 1p36.1-p34. It plays an important role in maintaining the glomerular ultrafiltration barrier and in reducing density. It can contribute to the initiation of albuminuria and to the alteration of the renal function in patients suffering from type 1 diabetes mellitus. It has a role in the endothelial dysfunction and in improving capillary permeability.18 In vivo studies showed that HSPG has several cellular functions and is essential in development, that is why it is involved in the regulation of signalling pathways, transforming growth factor β and fibroblast growth factor pathways.22 It helps maintain vascular homeostasis and can participate in the activation of FGF2, by stimulating endothelial growth and regeneration;23 it also plays a role in lipoprotein metabolism.24 Polymorphism is associated with diabetes mellitus and HSPG (Heparan sulfate proteoglycans).18
ACE (Angiotensin-Converting Enzyme) gene. Cytogenetic location: 17q23.3. ACE plays an important role in regulating blood pressure,25 and in the conversion of (inactive) angiotensin I decapeptide to (active) angiotensin II octapeptide.26 A role in pulmonary infections caused by coronaviruses is held by ACE ID polymorphism, which is consedered to be involved in the spread of SARS-CoV-19 among European populations.27 ACE ID polymorphism is associated with cardiovascular diseases, diabetes mellitus, diabetic nephropathy, aterosclerosis, heart diseases and stroke, high blood pressure, obesity, Alzheimer disease, cancer and Parkinson disease (DD genotype), while II genotype has also a strong effect in longevity.26 It was reported that ACE2 deficiency in the mouse, which encodes an essential regulating enzyme of the renin-angiotensin system (RAS) results in a very high susceptibility in the epithelial lesion-induced intestinal inflammation. RAS is involved in acute pulmonary insufficiency, cardiovascular functions and SARS infections.28 ID polymorphism in intron 16 of ACE1 is associated with modifications of ACE circular and tissue concentrations. SARS-CoV and SARS-CoV-2 can bind to target cells via ACE2 whihg facilitates quick viral replication, while in the lungs, the depletion of ACE2 from the cell membrane improves the damaging effects of Ang II.29
FTO (Fat mass and obesity-associated) gene. Cytogenetic location: 16q12,2. FTO gene plays a role in obesity; through the generation of new fat cells or adipocytes, a process called adipogenesis, this gene acts on the key processes occurring in the first stages of adipogenesis.30 It is also involved in nutrient detection and translation and growth regulation.31 FTO is associated with type 2 diabetes mellitus and cardiovascular disease,32 obesity, cancer, hypertension, Alzheimer disease and renal impairment33 and with MS.34
ADRB3 (Beta-Adrenergic Receptors 3) gene. Cytogenetic location: 8p11.23. ADRB3 gene has a role in lipid metabolism35 and in the regulation of the energy balance.36 The polymorphism of gene ADRB3, Trp64Arg, is associated with obesity, diabetes mellitus and hypertension,37 metabolic obesity38 and MS.39
UCP2 (uncoupling protein 2) gene. Cytogenetic location: 11q13.4. UCP2 is involved in thermogenesis, obesity, diabetes mellitus and atherosclerosis, as well as in the control of the reactive oxygen species derived from mitochondria (Vidal-Puig et al., 1997). It also plays a role in ageing, as a source of amino acid, as cellular response to starvation, as cellular response to glucose stimulus, as cellular response to insulin stimulus, in pregnancy, liver regeneration, negative regulation of apoptosis, negative regulation of insulin secretion,40 in lipid metabolism (Pecqueur et al., 2009). UCP2 was associated with obesity,41 type 2 diabetes mellitus.42 It plays a role in the metabolic syndrome and in the development of type 2 diabetes mellitus.43
UCP3 (uncoupling protein 3) gene. Genomic location: 11q13.4.44 UCP3 can participate in the modulation of the respiratory control of tissues,40 in thermogenesis,44 in the outward translocation of fatty acids from the mitochondrial matrix.45 UCP3 is associated with severe obesity46 and type 2 diabetes mellitus.47
FABP2 (Fatty acid binding protein 2) gene. Genomic location: 4q28-q31. FABP2 is involved in the improvement of the intracellular enzyme hypofunction caused by long-chain fatty acids. It acts as a free radical scavenger or it removes peroxidized acids from cells.48 FABP2 is probably involved in the lipoprotein and triglyceride synthesis and can help maintain functional energy homeostasis as a lipid sensor; it responsible for the modulation of cell growth and proliferation.49 IFABP can be involved in the absorption of fatty acids from the intestinal lumen and the transport of intestinal enterocytes to organs,50 in the intercellular absorption and transport of dietary long-chain fatty acids.51 FABP2 gene is associated with MS,52 with type 2 diabetes mellitus53 and high blood pressure.54
PLIN1 (Perilipin 1) gene. Cytogenetic location: 15q26.1. PLIN1 gene controls the storage and release of fats into adipocytes;55 it acts as a modulator of lipid metabolism in adipocytes.56 PLIN1 is associated with insulin-dependent diabetes mellitus (IDDM3),57 with obesity,58 cardiovascular disease59 and metabolic syndrome.60
PON1 (paraoxonase/arylesterase 1) gene. Cytogenetic location: 7q21.3. It is responsible for the hydrolysis of organophosphorus pesticides and toxic fumes acting on the nervous system; it plays a role in the innate immunity and healthy ageing. However, this mechanism is yet unknown.61 It participates in lipid peroxidation, detoxification of reactive molecules, modulation of endoplasmic reticulum stress and regulation of cell proliferation/apoptosis.62 PON1 gene is associated with coronary diseases,63 diabetes and amyotrophic lateral sclerosis,49 type 1 diabetes mellitus,64 insulin resistance and metabolic syndrome,65 type 2 diabetes mellitus and its complications,66 and high blood pressure.67
BHMT2 (Betaine homocysteine methyltransferase 2) gene. Cytogenetic location: 5q14.1.68 BHMT2 is involved in the regulation of homocysteine metabolism.40 It is associated with cardiovascular disease, osteoporosis, dementia and pregnancy complications,69 coronary artery disease, stroke and venous thrombosis.70
MTHFR (5,10-Methylenetetrahydrofolate reductatse) gene. MTHFR is located on chromosome 1p36.3.71 MTHFR enzyme plays an important role in the processing of amino acids and blocks of proteins.72 The epigenetic functions of the methyl group are the following: to protect DNA and RNA against the action of viruses, bacteria, heavy metals, solvents and other toxins in the environment; to lower histamin levels; to protect cell membranes.73 It participates in the formation and maturation of RBC (red blood cells), leucocytes (white blood cells) and in the production of thrombocytes.74 MTHFR is associated with cardiovascular diseases,75 HBP and mental retardation76 and MS.77 The American Diagnosis and Statistical Manual of Mental Disorders (DSM-5) contains a number of criteria for the classification of mental disorders: dementia (BHMT2 gene), mental retardation (MTHFR gene), degenerative diseases: Alzheimer, which is caused by genetic mutations in persons with a family history and Parkinson (ACE gene).78 It aimed to involve ACE/MTHFR genotypes in a wider algorithm of genes which influence CVD/diabetes, obesity, along with the gene which the literature considers as critical in Alzheimer disease–ApoE. ApoE4 poses a high risk of developing late-onset Alzheimer disease and cardiovascular diseases.79
IRS1 (insulin receptor substrate 1) gene. Cytogenetic location: 2q36.3. IRS1 is involved in insulin signaling, maintaining basic cell functions, such as: growth, survival, metabolism.80 IRS1 is associated with IR syndrome, atherosclerotic cardiovascular diseases associated with type 2 diabetes mellitus,81 HBP82 and MS.83
TRPM6 (Transient Receptor Potential Cation Channel Subfamily M Member 6) gene. Cytogenetic location: 9q21.13.84 TRPM6 has an important role in epithelial magnesium transport and the absorption of active magnesium in the gut and kidneys.49 TRPM6 is associated with metabolic disorders and associated chronic diseases such as oxidative stress, systemic inflammation, endothelial dysfunction, insulin resistance, HBP, type 2 diabetes mellitus and coronary disease,84 diabetes mellitus,85 gestational diabetes mellitus.86
MT1A (Metallothionein 1A) gene. Cytogenetic location: 16q13. Metallothioneins participate in the zinc and copper metabolisms and can be involved in neutralizing free radicals87 and protecting against reactive oxygen species.88 MT1A is associated with type 2 diabetes mellitus and CVD,89 HBP, MS and obesity.90
APOA5 (Apolipoprotein A5) gene. Cytogenetic location: 11q23.3. APOA5 participates in determining plasmatic levels of triglycerides in an age-independent manner.91 It is associated with obesity and metabolic syndrome,91 HBP93 and DM2.94
PEMT (Phosphatidylethanolamine N-Methyltransferase) gene. Cytogenetic location: 17p11.2. The CDP-choline pathway participates in obtaining choline from diet, through lipid metabolism. The PEMT pathway was shown to have a critical role in supplying PC during periods of starvation. PC, via PEMT, plays numerous physiological roles: cholin synthesis, hepatocyte membrane structure, bile secretion and very low-density lipoprotein secretion. The binding site in the region of PEMT promoter may increase the risk of liver steatosis an estrogen choline deficiency.95 PEMT is associated with liver disorders96 and with DMZ.97
ABCB4 (ATP Binding Cassette Subfamily B Member 4) gene. Cytogenetic location: 7q21.12. This gene plays a role in the metabolism and homeostasis of glucose;98 it protects hepatocytes against the damaging activity of bile salts,56 participates in recruiting phosphatidylcholine (PC), phosphatidylethanolamine (PE) and sphingomyelin molecules (SM); 40 in bile formation, as a transporter of bile salts, it mediates the ATP-dependent lipid efflux and excretes phosphatidylcholine and cholesterol in the presence of bile salts;99 it participates in the transport of phospholipids from hepatocytes into the bile.100 ABCB4 is associated with severe liver diseases,101 liver steatosis,102 non-alcoholic fatty liver disease and obesity.103
CHDH (choline dehydrogenase) gene. Cytogenetic location: 3p21.104 CHDH is involved in physiological processes and aterogenesis.105 It is associated with CVD,106 type 2 diabetes mellitus and obesity.107
FADS2 (Fatty acid desaturase 2) gene. Cytogenetic location: 11q12.2.108 FADS2 participates in fatty acid metabolism via its PPAR signalling pathways, alpha-linoleic acid (omega 3) and linoleic acid (omega 6).49 FADS2 is associated with type 2 diabetes mellitus, CAD, MS, myocardial infarction and dyslipidemia,109 and with MS.110
PCYT1A (Phosphate cytidylyltransferase 1, choline, alpha) gene. Cytogenetic location: 3q29. PCYT1A is involved in the normal functioning of fatty tissue and insulin action,111 and in the control of phosphatidylcholine synthesis.112 PCYT1A is associated with severe fatty liver, DM and steatosis.113
PCYT1B (Phosphate cytidylyltransferase 1, choline, beta) gene. Cytogenetic location: Xp22.11. PCYT1B participates in the control of phosphatidylcholine biosynthesis and in the suppression of the growth of calcium oxalate (CaOx) crystals.114 PCYT1B is associated with CVD, DM, HBP and MS.115
PNPLA3 (Patatin like phospholipase domain containing 3) gene. Cytogenetic location: 22q13.31.116 The normal function of PNPA3 provides instructions to a protein called adiponutrin, which can be found in fat cells (adipocytes) and liver cells (hepatocytes). The function of adiponutrin is not well known, but it is believed to help the regulation of adipocyte development, the fat production and decomposition (lipogenesis and lipolysis) in hepatocytes and adipocytes;8 it may be involved in the energy balance of adipocytes.116 PNPLA3 is associated with a whole range of non-alcoholic fatty liver lesions: steatohepatitits, cirrhosis and hepatocellular carcinoma (HCC), chronic viral hepatitis, alcoholic liver disease, haemochromatosis,117 obesity, liver steatosis,118 NAFLD, MS and type 2 diabetes mellitus.119
SCD (Stearoyl-CoA desaturase) gene. Cytogenetic location: 10q24.31.120 SCD1 plays an important role in the lipid metabolism.121 SCD is associated with CVD, obesity, non-insulin-dependent diabetes mellitus, HBP, neurological diseases, immune system disoders and cancer,121 type 2 diabetes mellitus and MS.122
SLC44A1 (Solute Carrier Family 44 Member 1) gene. Cytogenetic location: 9q31.2. SLC44A1 acts as a choline transporter in the central nervous system.123 It participates in membrane synthesis and myelin production.124 SLC44A1 is associated with liver steatosis, MS and type 2 diabetes mellitus.125
STAT3 (Signal Transducer And Activator Of Transcription 3) gene. Cytogenetic location: 17q21.2.126 The transcription factor STAT3 is constitutively active in many types of cancer, where it mediates important biological effects, including cell proliferation, differentiation, survival and angiogenesis. The N-terminal domain of STAT3 fulfills multiple functions, such as cooperative DNA binding, nuclear translocation.127 STAT3 gene is associated with fatty liver diseases,126 MS and abdominal obesity.128
UBE2E2 (Ubiquitin Conjugating Enzyme E2 E2) gene is located on chromosome 3p24.3.49 It participates in insulin secretion and synthesis. It is associated with type 2 diabetes mellitus129 and MS.130
ADAMTS9-AS2 (RNA gene ADAMTS9-Antisense RNA2) gene is located on chromosome 3p14.1.49 It is involved in the inhibition of the proliferation and migration of non-small cell lung cancer cells(NSCLC) and in renal cell carcinoma(RCC).131 It is associated with type 2 diabetes mellitus and obesity.132
KLHDC5 (Kelch Domain-Containing Protein 5) gene is located on chromosome 12p11.22. It plays a role in the microtubule dynamics during mitosis.49 It is associated with type 2 diabetes mellitus, obesity and metabolic syndrome.133
MEFV(Mediterranean fever sau Pyrin Innate Immunity Regulator) gene. It is located on chromosome 16p 13.3. 49 It is involved in the abnormal purine synthesis, to prevent the effects of suppression and inflammation. It is associated with MS.134
HFE (Homeostatic iron regulator) gene. It is located on chromosome 6p22.2.49 It is involved in the absorption of circulating iron via the regulation of the interaction of transferrin receptor with tranferrin.135 This gene is associated with type 2 diabetes mellitus and MS.136
PPARA (Protein Coding, Peroxisome Proliferator Activated Receptor Alpha) gene. It is located on chromosome 22q13.31.49 It is involved in the regulation of carbohydrate and lipid metabolism. This gene is associated with MS and type 2 diabetes mellitus.137
SLC16A1 (Solute carrier family 16 member 1) gene. It is located on chromosome 1p13.2.49 It participates in the energy (glucose) supply, particularly in the brain, which is not able to use fatty acids directly.138 This gene is associated with MS and obesity.139
Advanced NGx test uses the sequencing method, which allows several iterations (repeats) of the DNA sequence identifiction reaction, by which several genetic variations can be identified.4
Odds Ratio is a mathematical method to calculate the risk of disease conferred by genotypes.140
Software Jalview is a program for the 3D vizualisation of the secondary structure and the nucleic acid composition of a DNA sequence.5
MDR (Multifactor dimensionality reduction software). MDR is a method used to detect gene-environment interactions, based on the dimension reduction to one size. One challenge in human genetics is identifying polymorphisms or DNA sequence variations which present a high risk of disease. In order to detect genotypes interactions, predictor genotypes are effectivley reduced from N-dimensions to one dimension.141
The conclusion is that the clinical data, the biochemical and hematological laboratory investigations and the genetic variations of the 28 genes of interest: UCP2, UCP3, FABP2, PLIN1, PON1, FTO, ADRB3, BHMT2, MTHFR, IRS1, TRPM6, MT1A, APOA5, PEMT, ABCB4, CHDH, FADS2, PCYT1A, PCYT1B, PNPLA3, SCD, SCL44A1, STAT3, UBE2E2, ADAMTS9-AS2, KLHDC5, MEFV, MFE, INS, IGF2, ACE, TGF beta, HSPG Bam H1, sequenced by the Advanced NGx test and statistically interpreted by Jalview 2.8, MDR software version 3.0.2. programs, were correlated and they are associated with a high risk of developing metabolic syndrome.142-145
I wish to express my gratitude to all those people who provided qualified guidance and effective support in carrying out this study: Professor Tatiana Vassu-Dimov PhD, scientific coordinator of my doctoral thesis, Mr. Dănuț Gheorghe Cimponeriu, Professor at the Genetics Department of the Faculty of Biology, Bucharest University, Ms. Natalia Cucu Professor at the Genetics Department of the Faculty of Biology, Bucharest University, Ms. Ileana Stoica Professor at the Genetics Department of the Faculty of Biology, Bucharest University, Ms. Alexandra Simon Gruiță Professor at the Genetics Department of the Faculty of Biology, Bucharest University for allowing me to use the facilities of the Human Genetics Laboratory in conducting the genetic tests, Dr. Crăciun Anne Marie, who allowed me to use the biological samples of “N. Paulescu” Institute for Diabetes, Nutrition and Metabolic Diseases, Bucharest, and Dr. Popescu-Guinea Gelu, head of the Diabetes, Nutrition and Metabolic Diseases Section within the Internal Diseases Department of Giurgiu County Emergency Hospital, who indicated to me the clinical criteria for the selection of patients with metabolic syndrome, obesity and diabetes mellitus, whose biological samples were investigated in the Clinical Bacteriology Laboratory of Giurgiu County Emergency Hospital under the guidance of Dr. Matefi Felicia, head of the laboratory, and Dr. Mihai Petre, Manager of Giurgiu County Emergency Hospital, who authorized my internship on a volunteer basis (Volunteering Contract no. 2/20.01.2014), thus enabling me to gather the biological material included in this study.
Authors declare that there is no conflict of interest.

©2020 Alina. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.