MOJ
eISSN: 2374-6920


Research Article Volume 7 Issue 4
1Department of Natural Sciences, LaGuardia Community College, New York, 1110, USA
2Division of Nephrology, Department of Medicine, State University of New York, Downstate Medical Center, Brooklyn, New York, 11203, USA
3Department of Chemistry, Lehman College of the City University of New York, Bronx, New York, 10468, USA
4Biological Mass Spectrometry Core Facility, University of Colorado Denver, Aurora, CO 80045, USA
5Pathology Department, Albert Einstein College of Medicine, Bronx, NY, 10461
Correspondence: Cristina C Clement, Department of Pathology, Albert Einstein College of Medicine, Bronx, NY, 10461, USA, and Department of Chemistry, Lehman College of the City University of New York, Bronx, New York, 10468, USA, Tel 718-430-3468
Received: August 13, 2018 | Published: August 22, 2018
Citation: Gonzalez J, Babinska A, Ewul ELV, et al. Development of a label-free nanolc/ms/ms assay for monitoring the changes in the proteomic landscape of thrombin?activated human platelets. MOJ Proteomics Bioinform. 2018;7(4):232-255. DOI: 10.15406/mojpb.2018.07.00242
One top research assay used to validate the efficacy of pharmacological agents used in antiplatelets aggregation therapy is mass spectrometry coupled with high throughput profiling of changes in the proteomic expression in the activated platelets exposed to inhibitors or agonists of platelets signaling. Herein we present the development of a new, bottom‒up proteomics platform that enabled monitoring of changes in protein expression profiles of healthy human platelets, ex‒vivo activated with thrombin, using as reference the proteome of resting platelets. The assay employed the platelets cryogenic lysis and protein solubilization under denaturant conditions, enabling the extraction of key membrane‒bound proteins, in addition to the cytosolic and organelles‒derived proteins secreted in microparticles, and found in the platelets releasates. Nano LC‒ESI/MS/MS sequencing of tryptic/Glu‒C/Lys‒C peptides from the “in solution” one step digestion of the whole platelets proteomes (2x108 platelets/patient sample) was performed on a Q‒Exactive quadrupole orbitrap mass spectrometer coupled with label free quantification (LFQ) analysis. The changes in proteomic landscapes were qualitatively and quantitatively analyzed using a combination of systems biology and bioinformatics approaches empowered by the ingenuity pathway analysis (IPA), and the screening of identified proteins and genes entities against curated databases containing platelets proteomes, including the “Adhesome”, “Exocarta”, “Reactome” and “Platelet Web”. The label‒free global proteomics analysis retrieved a total of 924 proteins (FDR <1.3% for proteins and <0.8% for peptides) out of which 330 were shared between resting and thrombin‒activated platelets. About 50% of the total proteome of thrombin‒activated platelets was represented by up‒regulated and previously validated protein biomarkers, involved in the pathways mediating the protease and thrombin activated receptor (PARs), integrin signaling, the actin and rhoA linked to cytoskeleton signaling, and activated kinases pathways regulating the cellular motility, aggregation, procoagulation and degranulation. In conclusion, the systems biology approaches validated our newly developed LFQ proteomic assay as a reliable tool for monitoring the changes in protein expression profiles in thrombin‒activated platelets, and further advocate for employment of these approaches in assessing the efficacy of antiplatelets drugs (inhibitors of platelets aggregation) during the prevention and treatment of cardiovascular and cerebrovascular diseases.
Keywords: whole platelet proteomics, nano LC‒ESI/MS/MS, LFQ analysis, systems biology, bioinformatics, platelet web
Platelets are small anucleated blood particles derived from megakaryocytes in the bone marrow, and they play a key role in the control of bleeding and hemostasis.1‒10 Platelets need to be activated to perform their functions, a process mediated by many physiological activators. The best known and characterized activators at the level of signaling transduction mechanisms are: thrombin, arachidonic acid and its derivatives (thromboxane A2), collagen, and the adenosine nucleotides (ADP and ATP).2‒8 Activated platelets undergo vast cytoskeleton, organelles, and secretory protein reorganization accompanied by many posttranslational modifications (PTM).5‒14 Due to their anucleate nature, platelets have limited protein synthesis, and therefore it is expected that most of the changes encountered at the level of the proteome under different pathophysiological conditions will be determined mostly by changes in protein expression (translational and post‒translational regulated), at the level of PTM, proteolysis, or secretion during platelets degranulation processes associated with their activation.11‒15 It is well documented that selected platelet messages are translated into proteins after activation, regulating the inflammatory and hemostatic responses of the platelet.2‒8 Platelets can be affected by many physiological conditions during blood circulation that can ultimately lead to vascular complications initiated by thrombus formation.12‒16 During activation, release of platelet microparticles can generate various pathophysiological effects, such as initiation and exacerbation of stroke.16,17 One of the most advanced studies applied to the research of platelets biology under different activation states is the mass spectrometry coupled with profiling of protein changes applied to both resting and activated whole platelet proteomes, or to platelet subproteomes, including the platelet granules (alpha and dense), membrane systems, lipid rafts, and enriched phosphoproteome. Research conducted in the last five years acknowledges the identification of 5000‒5500 expressed proteins in human platelets, a proteome that is highly similar between different healthy individuals.10‒21 Recent emerging analytical technologies coupling the fractionation of the cellular proteome with the multidimensional nano LC/ESI/MS/MS sequencing, together with systems biology approaches empowered by bioinformatics analysis and data mining enabled a revolution in platelet proteomics that led to the development of new, mass spectrometric‒based assays for enhanced research of platelet pathophysiology.15‒21 Consequently, the modern proteomics approaches can reveal: (1) targeted and untargeted quantitative changes in the abundances of thousands of proteins; (2) post‒translational modifications; (3) protein‒protein interactions networks; (4) subcellular localization, i.e. membrane‒bound proteins, or secreted proteome in the microvesicles, exosomes, microparticles found in platelets releasates.13‒16 Using highly sensitive and highly specific mass spectrometers (providing mass accuracy and resolution), and proteome fractionations assays involving denaturant, and reducing one‒dimensional or two‒dimensional protein electrophoretic systems (1‒DE and 2‒DE, respectively) followed by protein digestions with multiple enzymes (trypsin, LysC, Glu‒C and Asp‒N, among others), and further fractionations of peptides by means of orthogonal chromatographic systems, thousands of proteoforms can be characterized from highly pure fractions consisting of only 2x108 platelets, purified from small volumes of blood, usually in the range of few milliliters.20‒28 Consequently, the future advances in the mass spectrometric profiling of protein expression and their corresponding highly‒regulated PTM are expected to allow platelet proteomics to be implemented in many research and clinical laboratories as a reliable tool for characterizing the molecular and cellular pathways that affect platelet homeostasis in healthy and disease states. Many original research articles and recent reviews highlighted the wide area of research dedicated to label or label‒free, targeted or untargeted proteomics profiling of changes in the platelets purified from patients experiencing cardiovascular, inflammatory, and bleeding disorders.27‒50
Among the most cited research studies are the pharmacoproteomic assays that rely on applications of both qualitative and quantitative protein expression profiling of activated platelets induced by various physiological agonists, such as thrombin, collagen, arachidonic acid, and ADP. Such assays serve to monitor and quantify the dynamic changes in the whole platelet proteomic landscape in the presence of different agonists or antagonists of platelets aggregation, among which the effects of aspirin +/‒ clopidogrel and of natural products, like curdione and sarpogrelate are the best known to date.18,51 In addition, these pharmacoproteomic approaches were successfully employed to study the effects of antiplatelet therapy for finding the best management of cardiovascular diseases (CVDs) and other inflammatory diseases.20,25,32,40‒42,48‒58 It is very well known that platelets play a pivotal role in the pathophysiological mechanisms underlying many CVDs, especially acute coronary syndromes (ACSs), since platelets are the key players in thrombus formation after atheroma plaque rupture.29,31,35. Accordingly, many molecules, lipids and proteins, especially thrombin and thrombin‒activated protease (PAR) receptors (PAR1 and PAR4), involved in platelet activation and aggregation, are primary targets for treatment of ACSs. Therefore, the search for drugs that inhibit platelet activation was common to all these studies, and the search for protein profiles indicative of the response to a selected drug and dose was facilitated using proteomic technologies. Consequently, the recent advances in platelet proteomics applied to the study of CVDs focus primarily on sample preparation, subcellular fractionation, and bioinformatics requirements for proteome analysis of platelets derived from clinical samples.30‒41,52‒58 To better understand the platelets’ response to thrombin activation and its subsequent engagement of PAR receptors under clinical circumstances, especially when potent thrombin inhibitors can be used to treat the ACS syndrome, we developed and optimized a bottom‒up, qualitative and quantitative label‒free proteomic research study of human platelets isolated from healthy donors and ex‒vivo activated with thrombin, using as reference the proteome of resting platelets. The label‒free global proteomics analysis retrieved a total of 924 proteins, representing about 17% from the total proteins reported to date for platelets proteomes.30‒41,50. About 50% of the total proteome of thrombin‒activated platelets was represented by up‒regulated previously validated protein biomarkers involved in the pathways mediating the protease and thrombin activated receptor (PARs) and integrin signaling, the actin and rhoA linked to cytoskeleton signaling, and activated kinases pathways regulating the cellular motility, aggregation, procoagulation, and degranulation. These results validated our new simple and expensive less LFQ proteomic assay as a reliable tool for monitoring the changes in the protein expression profiles in thrombin‒activated platelets that can be further implemented in the clinical environment where the research on discovery of antiplatelets drugs (inhibitors of platelets aggregation) is a key factor for advancing the prevention and treatment of cardiovascular and cerebrovascular diseases.
Human subjects and blood collection
Venous blood was collected from three healthy volunteers, who did not use any medication in the past 10days, as described in the previous published protocols.30‒35 The collection of blood from the volunteers was performed conform with the declaration of Helsinki and this part of the study was approved by the ethics review board of the Department of Medicine at SUNY, Downstate Medical Center, Brooklyn, New York, USA. Blood for MS‒based proteomics analysis was collected with an open system, anticoagulated with 3.2% trisodium citrate. Platelet‒rich plasma (PRP) was prepared by centrifugation of whole blood at 165xg for 10min at room temperature, no brake, as described in the previous published protocols.30‒35
Reagents
Trifluoroacetic acid, acetonitrile, acetic acid, formic acid, iodacetamide and methanol were purchased from Fisher Scientific (Pittsburgh, PA). Urea, dithiothreitol (DTT), TCEP‒HCL (Tris (2‒carboxyethyl) phosphine hydrochloride) iodoacetamide, ammonium bicarbonate, NaCl, KCl, KH2PO4, H3PO4 and NH4CO3 were purchased from SIGMA (St. Louis, MO, USA) and from Thermo Fisher Scientific (Waltham, MA USA). Complete Proteinase inhibitor cocktail was also purchased from Roche. Porcine trypsin (20ug, specific activity > 5,000units/mg seq. grade modified), Lys‒C (sequencing grade, 10ug) and Glu‒C, sequencing grade (10ug) were purchased from Promega (Madison, WI). Human α‒Thrombin (Factor IIa) (catalog#: HT 1002a, Size: 1000units) was from Enzyme Research Laboratories (South Bend, IN, USA). Pierce C‒18spin columns (cat#89870) were from Thermoscientific (Rockford, IL, USA).
Platelets purification and ex‒vivo activation with thrombin for proteomic analysis
PRP were subjected to the whole platelets purification as described previously by other research groups.30‒35 Then, platelets were pelleted at 750xg for 10min and twice washed with the Tyrode’s Buffer containing 134mM NaCl, 12mM NaHCO3, 2.9mM KCl, 0.34mM Na2HPO4, 1mM MgCl2, and 10mM Hepes (pH 7.4). Platelet count was measured with a Beckman Coulter LH780 system (Beckman Coulter, Inc., Brea, CA). The flow cytometric assessment showed that the platelet samples contained 99.15–99.35% CD61 positive cells (platelets), the rest being represented by leukocyte population (CD45 positive), a purity which was within the same range with other published research studies on whole platelets proteome. To obtain thrombin activated platelets, wash platelets were resuspended in 0.5mL of washing buffer with 1.8mM CaCl2 (to have 2x108 platelets/ml) and 1 IU of human alpha‒thrombin for each sample derived from three healthy volunteers.
Platelets lysis and proteins extraction
The washed platelets enriched fraction from three individual healthy donors was aliquoted in half for obtaining the whole proteome of “resting” and “thrombin‒activated” platelets, such that 2x108 platelets/ml was processed for the proteomics assay. The assay employed the platelets cryogenic lysis (5 cycles of freezing in liquid nitrogen and thawing at 37oC) followed by protein solubilization with a 50mM ammonium bicarbonate buffer (pH 8.5) containing 8M urea, without fractionation. The extraction of platelets proteome was performed by centrifugation at 18,000rpm for 30min, at 4oC in bench microcentrifuge. These cryogenic lysis and protein extraction under denaturant conditions enabled the extraction of key membrane‒bound proteins in addition to the cytosolic and organelles‒derived proteins secreted in the microparticles and found in the platelets releasates, as shown by our new developed bottom‒up proteomics analysis.
Sample preparation for mass spectrometry
The total extracted proteome from equal aliquots, (i.e.2x108 platelets/sample) was reduced in solution with 30mM TCEP for 40minutes at room temperature, followed by alkylation with 120mM iodoacetamide, for 55minutes, in the dark and at room temperature, in a buffer with 50mM ammonium bicarbonate (ABC) and 8M urea. The digestion was performed using combination of LysC/trypsin and Glu‒C enzymes, and the 1:25 protein: enzyme ratios, after urea was diluted to 2M final concentration with 50mM ABC buffer. The digestion was performed at 37oC for 16hours. The enzyme digestions were stopped with formic acid and acetonitrile (5% and 10%, respectively, final concentrations). The samples were further spun for 30minutes at 14,000xrpm in the top bench microcentrifuge. The supernatants containing the extracted peptides were evaporated in a Speed Vac, followed by desalting and concentration with C18 Zip Tip (Millipore) for further analysis using nano LC–ESI–MS/MS.
Nano LC–ESI–MS/MS analysis of tryptic/Glu‒C/Lys‒C peptides digests
The tryptic/Glu‒C/Lys‒C peptides digests from the whole platelets proteomes were analyzed on a Q Exactive quadrupole orbitrap mass spectrometer coupled to an Easy nLC 1000 UHPLC (Thermo Fisher Scientific) through a nano electrospray ion source. Peptides were separated on a C18 analytical column (100µm internal diameter, x20cm length) packed with 2.7µm Phenomenex Cortecs particles, as described in another published protocol (60). The solution of peptides was equilibrated with 3µL 5% acetonitrile 0.1% formic acid, before being separated using optimized 90, 120 or 180min linear gradients employing 2% to 32% acetonitrile in a buffer with 0.1% formic acid, at a flow rate of 400nL/min. The LC mobile phase solvents and the sample dilutions contained 0.1% formic acid in water (Buffer A) and 0.1% formic acid in 100% acetonitrile (Buffer B) (Optima™ LC/MS, Fisher Scientific, Pittsburgh, PA). Data acquisition was performed using the instrument provided Xcalibur™ (version 3.0) software. The mass spectrometer was operated in the positive ion ionization and data–dependent acquisition (DDA) mode. The full MS scans were obtained with a range of m/z 300 to 2000, at a mass resolution of 120,000 at m/z 200, and a target value of 1.00E+06 with the maximum injection time of 50ms. HCD collision was performed on the 15 most significant peaks, and tandem mass spectra were acquired at a mass resolution of 15,000 at m/z 200 and a target value of 1.00E+05 with the maximum injection time of 110ms. The dynamic exclusion time was 20s. The normalized collision energy was optimized for 32‒34% as described in a previous published protocol.59
Peptide identification and validation
Raw files from each technical and biological replicate were filtered, de novo sequenced and assigned with protein ID using Peaks 8.5 software (Bioinformatics Solutions, Waterloo, Canada), by searching against the human (Homo Sapiens) Swiss Prot database (January 2017; 158,154 entries) using protocols published elsewhere.59‒61 The following search parameters were applied for LFQ analysis: trypsin/LysC and Glu‒C restrictions for enzymes and one allowed missed cleaved at one peptide end. The parent mass tolerance was set to 15ppm using monoisotopic mass, and a fragment ion mass tolerance was set to 0.06 Da. Carbamidomethyl cysteine (+57.0215 on C) was specified in PEAKS as a fixed modification. Methionine, lysine, proline, arginine, cysteine and asparagine oxidations (+15.99 on CKMNPR), deamidation of asparagine and glutamine (NQ‒0.98) and pyro‒Glu from glutamine (Q‒18.01 N‒term) were set as variable modifications. Data were validated using the FDR method built in PEAKS 8.5,40,41 and protein identifications were accepted with a confidence score (‒10lgP)>18 for peptides and (‒10lgP)>15 for proteins; a minimum of 1 peptide per protein after data were filtered for less than 1.5 % FDR for peptides and less than 0.9% FDR for proteins identifications. An independent spectra filtering and validation of the MS/MS‒spectra and identified peptides and proteins was performed using Scaffold (version Scaffold_4.8.7, Proteome Software Inc., Portland, OR). The Scaffold built in option “MuDPIT” was used to combine multiple files from technical/biological replicates for each sample defining the “resting” and “thrombin‒activated” platelets samples.
Criteria for protein identification in proteome software
Scaffold was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Peptide Prophet algorithm,59 with Scaffold delta‒mass correction. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 1 identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm.59 Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Peptide identifications were set at greater than 95.0% probability by the Peptide Prophet algorithm with Scaffold delta‒mass correction. Protein identifications were set at greater than 95.0% probability and contained at least 1 identified peptide. These parameters ensured a final FDR<1.3% for proteins and <0.8% for peptides. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Label‒free quantitative (LFQ) methods (precursor intensity (MS1) and spectral counts (MS2)) were used to analyze and contrast the proteomic profiles “resting” and “thrombin‒activated” human platelets
LFQ analysis based on the precursor ion (MS1) intensity was performed using the quantification algorithm supported by the Quantitative PEAKS Q module (Bioinformatics Solution Inc., version 8.5) using an optimized protocol published elsewhere.59‒61 Briefly, the data were filtered, smoothed, and aligned in retention time, followed by feature detection based on peak volume and isotopic clustering using the algorithm of PEAKS 8.5. MS/MS spectra were then extracted using the same software and used to search against the target‒decoy database containing all Swiss Prot entries for the Homo Sapiens (January 2017; 158,154 entries) as described above. The relative protein abundances were exported for all proteins representing each biological sample, “resting” vs “thrombin‒activated” platelets, after normalization of the corresponding averaged areas (abundances) with respect to the total ion current (TIC). Only proteins which passed a selected significance statistical threshold (ANOVA, p<0.05 and FDR <1% for protein and peptide expression) across all samples were used to perform the quantitative analysis. A complementary LFQ analysis using the spectral counts (specifically the normalized weighted spectral counts) intensity, i.e. based on MS/MS or MS2 analysis, was employed for the same set of platelets proteomic samples using the normalized weighted spectrum count option provided in “Scaffold” (version 4.8.7) and “per SPECtives” (version 2.0.6, Proteome Software Inc., Portland, OR). The Scaffold rescaled the protein ratios values using a log2 transformation, such that positive values reflected fold increases while the negative values reflected fold decreases. The final quantitative profile associated with each sample were statistically evaluated using the statistical significance threshold (p<0.05, by Fisher exact test). The matching “mzIdent ML” files generated in Scaffold for each of the “resting” and “thrombin‒activated” platelets biological sample were imported in “per SPECtives” software. The proteins were clustered in representative heat maps using a single linkage method and a ranked‒based Euclidean distance‒metric algorithm together with the weighted spectra counts for protein abundances. The Fisher’s test followed by standard Benjamini‒Hochberg correction was used to determine the statistical significance for the fold changes (p‒values of <0.05 were considered significant).
Gene ontology (GO), molecular and cellular pathways enrichment and systems biology analysis of platelets proteome
Networks, functional analyses, biochemical and cellular pathways were generated using the ingenuity pathway analysis (IPA; Ingenuity Systems, Redwood City, CA, USA) and the list of proteins extracted from LFQ analyses, using a methodology developed by us for other proteomics data set and published elsewhere.59 Specifically, the experimentally determined protein ratios (from MS1 integrated areas and/or MS2 normalized weighted spectral counts) were used to calculate the experimental fold changes by rescaling their values using a log2 transformation, such that positive values reflected fold increases while the negative values reflected fold decreases. For network generation, datasets containing gene identifiers (gene symbols) for the “resting” and “thrombin‒activated” platelets were uploaded into the IPA application together with their rescaled log2 transformation of protein’s area ratios. These molecules were overlaid onto a global molecular network contained in the Ingenuity Knowledge Base. The networks were generated based on their connectivity index using the built‒in IPA algorithm. The probability of having a relationship between each IPA indexed biological function and the experimentally determined genes were calculated by a right‒tailed Fisher’s exact test. The level of significance was set to a p‒value of <0.05. Accordingly, the IPA analysis identified the molecular and cellular pathways from the IPA library of canonical pathways that were most significant to the dataset (‒log (p value)>2.0). For the quantitative analysis of the expression profiles, IPA assigned the “z‒score” function to all eligible canonical and cellular pathways (where a “z<‒2 represent significant down‒regulation while a z>2.0 represent a significant up‒regulation of the selected pathway). The specific “resting” and “thrombin‒activated” associated platelets proteins biomarkers together with all the cellular pathways were assigned to each proteome data set using the following databases: Adhesome (literature‒based protein‒protein interaction network) (http://www.adhesome.org/); Exocarta (exosome proteins, RNA and lipids database E) (http://exocarta.org/); Reactome (free, open‒source, curated and peer‒reviewed biological pathways database (https://reactome.org/); Platelet Biology/Systems Biology Workbench (protein‒protein interactions and phosphorylations in human platelets database) (http://plateletweb.bioapps.biozentrum.uni‒wuerzburg.de/plateletweb.php).
Platelets play an important role in the hemostatic machinery, by responding rapidly to damage of the vascular wall. This response is mediated through several signaling pathways acting downstream of receptors for von Willebrand Factor (vWF), thrombin (the PAR receptors) and collagen.1‒20 Autocrine stimulatory factors such as ADP and thromboxane are important for stimulating further adhesion to fibrinogen, vWF and collagen (through integrin receptors αIIbβ3, α2β1) and for activating circulating platelets.5‒10 Platelets play a pivotal role in the pathophysiological mechanisms underlying a wide family of CVDs, especially acute coronary syndromes (ACSs), as key players in thrombus formation after atheroma plaque rupture.20‒45 To better understand the platelets response to the thrombin activation and its subsequent engagement of PAR receptors under clinical circumstances, when potent thrombin inhibitors can be used to treat the ACS syndrome, we conducted a qualitative and quantitative label‒free proteomic research study of human platelets isolated from healthy donors and ex‒vivo activated with thrombin, using as reference the proteome of resting platelets. Our main goal was to develop and optimize a bottom‒up label‒free proteomics assay which would use a small number of highly purified platelets (about 2x108 platelets/patient sample) from the few milliliters of blood from healthy donors (maximum 8ml). We used the clinical samples of purified platelets from three healthy donors, who were not undergoing antiplatelet or non‒steroidal anti‒inflammatory (NSAID) drug therapy. Each blood sample was split in half and used for proteome extractions from equal number of “resting” and ex‒vivo “thrombin‒activated” platelets. Given the intensive work required to fractionate the proteome and the expensive analytical mass spectrometer tools required to acquire the proteomic raw files, we prioritized the development of a simple method for proteome extraction avoiding multiple fractionation steps, but still suitable for retrieving the major known protein markers of thrombin‒activated platelets, mediating the cytoskeleton and integrin signaling and the degranulation pathways during the aggregation, as extensively reported by other proteomics studies,10‒45 the final goal being the implementation of a such proteomic assay for the basic and research clinical labs involved in the management of ACS using antiplatelet therapy.
Development of a workflow for implementing the label‒free pharmacoproteomics assay designed to monitor the changes in the proteomics profiles of human “resting” platelets from healthy donors in comparison with the ex‒vivo “thrombin activated” platelets
The outline for the workflow employed for the platelets proteomic analysis is presented in Figure 1. The thrombin activated platelets are expected to trigger intracellularly signaling pathways mediated by PAR‒1 and PAR‒4 receptors.1‒10 A new method for lysis the platelets in one single step, without prior separation by other analytical methods followed by the “in solution” digestion with three enzymes (trypsin/LysC/Glu‒C) enabled the extraction of peptides from the total platelet proteome, including the membrane‒derived and microvesicles contained in the releasates, that were amenable for nano LC/MS/MS sequencing on a Q Exactive quadrupole orbitrap mass spectrometer. The bottom‒up label‒free proteomics analysis employed the label free quantification (LFQ) module provided by PEAKS 8.5 (Bioinformatics Solutions Inc.) on proteomic samples processed from equal number of platelets (2x108cells/treatment). The changes in the protein expression profiles were complemented by an additional analysis involving the Scaffold and per SPECtives software (Proteome Software Inc.) where the protein abundances are reported by LFQ analysis employing spectra counts. The platelets proteomes were further annotated for their gene ontology (GO), molecular functions, cellular pathways and protein‒protein interactions networks using a comprehensive bioinformatics and system biology approaches employing the Ingenuity Pathways Analysis (IPA), “Exocarta‒Funrich”, “Reactome”, “Adhesome” and “Platelet Web/Systems Biology” webservers.
The thrombin activated platelets are expected to trigger intracellularly signaling pathways mediated by PAR‒1 and PAR‒4 receptors.13,15 To achieve a high sequence coverage for the proteins involved in the PAR‒1 and PAR‒4 signaling transduction and involved in platelets aggregation events, we developed and optimized a method for platelets lysis in absence of any detergent, i.e. a cryogenic lysis in liquid nitrogen followed by protein solubilization under denaturant conditions (in 50mM ammonium bicarbonate buffer, pH 8.5 with 8M urea) Figure 1. Equal aliquots of start‒up platelets (2x10^8 cells/sample) were used for the “resting” and the “thrombin‒activated” platelets. Three biological replicates were subjected for the bottom‒up analysis, each run in technical dupliactes. The “in solution” digestion with three enzymes (trypsin/LysC/Glu‒C), enabled the extraction of peptides from the total platelet proteome, including the membrane‒derived and microvesicles contained in the releasates, as reported previously by other studies on platelets proteomics.20‒30 The extracted peptides were subjected to nano LC/MS/MS sequencing on a Q Exactive quadrupole orbitrap mass spectrometer, as summarized in the protocol shown in Figure 1. The label‒free proteomics analysis employed the label free quantification (LFQ) module provided by PEAKS 8.5 (Bioinformatics Solutions Inc.), In addition, the changes in the protein expression profiles were complemented by an additional analysis involving the Scaffold and per SPECtives software (Proteome Software Inc.). The platelets proteomes were further annotated for their gene ontology (GO), molecular functions, cellular pathways and protein‒protein interactions networks using a comprehensive bioinformatics and system biology approaches employing the Ingenuity Pathways Analysis (IPA), “Exocarta‒Funrich”, “Reactome”, “Adhesome” and “Platelet Web/Systems Biology” webservers Figure 1.

Figure 1 Workflow displaying the strategy employed for the label‒free pharmacoproteomics approach designed to monitor the changes in the proteomics profiles of human “resting” platelets from healthy donors in comparison with the ex‒vivo “thrombin activated” platelets.
Nano LC/MS/MS analysis of tryptic peptides derived from the whole proteomic analysis of “resting” and “thrombin‒activated” platelets
Mass spectrometric analysis of MS1 and MS2 data acquired on a Q Exactive quadrupole orbitrap mass spectrometer employing the HCD positive ionization mode. As shown in Figure 2. Mass spectrometric analysis of MS1 and MS2 data acquired on a Q Exactive quadrupole orbitrap mass spectrometer employing the HCD positive ionization mode. (A and B): Representative 2D heat maps (shown as retention time (RT) vs m/z), corresponding to the total number of features extracted from the filtered MS scans, having aligned RT, m/z, mass error (ppm), charge (z) and average area (in purple color), are displayed together with the total number of MS/MS spectra assigned to peptide sequences (blue color). The 2D heat maps represent the proteomic output from LFQ analysis of one (out of three) representative biological samples corresponding to the “resting” and “thrombin‒activated” platelets. On average, between 3600‒10,000 peptides sequences were successfully assigned by de novo sequencing coupled with LFQ and PEAKS ID analysis, using PEAKS 8.5 software, for each biological sample comprising of “resting” or “thrombin‒activated” platelets. (C‒D) 3D (upper panel) and 2D (lower panel) intensities chromatogram views (RT vs (m/z) vs MS1/MS2 (“total ion current” TIC)) of nano LC/MS1/MS2 data for the proteomic and LFQ analysis of one representative biological sample recorded for the “resting” and “thrombin‒activated” platelets.

Figure 2 nano LC/MS/MS analysis of tryptic peptides derived from the whole proteomic analysis of “resting” and “thrombin‒activated” platelets.
The representative 2D heat maps (retention time (RT) vs m/z) show a high overlap (about 85‒90%) between the total number of features extracted from the filtered MS scans, characterized by aligned RT, m/z, mass error (ppm), charge (z) and average area (purple color) and the total number of MS/MS spectra assigned to peptide sequences (blue color). The 2D heat maps represent the proteomic output from LFQ analysis of one (out of three) representative biological samples corresponding to the “resting” and “thrombin‒activated” platelets Figure 2). On average, between 3600‒10,000 peptides sequences were successfully assigned to protein IDs using the de novo sequencing coupled with LFQ and PEAKS ID analysis using PEAKS 8.5 software, for each biological sample comprising of “resting” and “thrombin‒activated” platelets (see Figure 3 for statistics of PEAKS analysis). Representative qualitative results of proteomic output for one selected biological sample (thrombin‒activated platelets), performed with PEAKS 8.5, after de novo sequencing and PEAKS ID coupled with LFQ analysis. (A) The PEAKS built‒in false discovery rate (FDR=0.1%) is displayed for the identified peptide‒spectrum matches (PSM). (B) The distribution of PEAKS peptide score (calculated as‒10lgP) (panel (a) in B) is presented with the PEAKS scatterplot of peptide score vs the precursor mass error (ppm) (panel (b) in B). The decoys are displayed in brown while the target hits are displayed in blue (in B). Peptides having PEAKS peptide score “‒10lgP” of 15 or 18 and higher were confidently assigned to proteins with PEAKS ID and characterized by FDR<0.8%. The complete list of proteins and details of mass spectrometry analysis for each of the proteomes of “resting “and “thrombin‒activated” platelets are provided in the Supplementary Table 1. Nano UPLC gradients for fractionating the tryptic peptides were developed for 90, 120 and 180minutes, as displayed in the Figure 2C & Figure 2D and described in detail in the method section. The gradients employing 120 and 180minutes time for fractionation were the best suited to gain more than 3500peptides with assigned sequence, after MS/MS analysis (see Figure 3 for data analysis).

Figure 3 Label‒free qualitative analysis and protein identification for the proteomes from “resting” and “thrombin‒activated” platelets.
Label‒free qualitative and quantitative analysis of protein identification for the proteomes from “resting” and “thrombin‒activated” platelets
The analysis of MS1 and MS/MS data provided by the mass spectrometry collected raw files were analyzed with the “de novo” sequencing algorithm coupled with LFQ and PEAKS ID modules employed in PEAKS 8.5 software (Bioinformatics Solutions, Inc, Waterloo, Canada). Representative qualitative results of proteomic output for one selected biological sample (i.e. the “thrombin‒activated” platelets) are shown in Figure 3. The PEAKS built‒in false discovery rate algorithm was used to assess the quality of the assigned peptide sequences and to validate the MS/MS features with assigned sequence. As shown in the Figure 3A the calculated FDR was 0.1% for the peptides‒spectrum matches (PSM) while the distribution of PEAKS peptide score (calculated as‒10lgP) (panel (a) in B) are showing that peptides having PEAKS peptide score “‒10lgP” of 18 or 20 and higher could be confidently assigned to proteins with PEAKS ID. On average the calculated peptides FDR was <0.8%. The complete list of proteins and details of mass spectrometry analysis for each replicate proteome of “resting “and “thrombin‒activated” platelets, are provided in the Supplementary Table 1. The canonical pathway is displayed by IPA using the experimentally determined protein ratios (i.e. “activated”/”resting” platelets) quantified using ≥1 peptides/protein, and their associated fold changes, shown in the table as “Experimental Log ratios”. 52 proteins identified in the proteome of “thrombin‒activated” platelets are matched with the integrin signaling pathway by the knowledge database in IPA. The IPA rescaled the protein ratios values using a log2 transformation, such that positive values reflected fold increases (shown as “Up”) while the negative values reflected fold decreases (shown as “Down”). The integrin signaling pathway the assigned “activation z‒score” of “+5.28” (see details in Supplementary Table 4).
The validation of mass spectrometric MS1 and MS/MS (i.e. MS2) data was complemented by the statistical analysis of the proteomic output for each biological replicate using the PEAKS 8.5 software, after de novo sequencing and PEAKS ID coupled with LFQ analysis. As shown in Figure 4A, PEAKS software provides Table 1 containing the statistics for the total number (#) of MS and #MS/MS scans. This table is further complemented with the results for the filtration parameters, such as the peptide score, “A” score (A>20 assigned confident PTM), protein score, #unique peptides/protein, and de novo ALC score, presented in Table 2. The canonical pathway is displayed by IPA using the experimentally determined protein ratios (i.e. “activated”/”resting” platelets) quantified using ≥1 peptides/protein, and their associated fold changes, shown in the table as “Experimental Log ratios”. 47 proteins identified in the proteome of “thrombin‒activated” platelets are matched with the actin cytoskeleton signaling pathway by the knowledge database in IPA. The IPA rescaled the protein ratios values using a log2 transformation, such that positive values reflected fold increases (shown as “Up”) while the negative values reflected fold decreases (shown as “Down”). The cytoskeleton signaling pathway has an assigned “activation z‒score” of “+5.94” (see details in Supplementary Table 4).

Figure 4 Label‒free qualitative analysis of proteomes from “resting” and “thrombin‒activated” platelets.
The statistics for the filtered mass spectrometric results are displayed in Table 3 (panel A) from Figure 4B and includes the FDR for PSM, peptides and proteins, in this case for a biological replicate representing the “thrombin‒activated” platelets. Statistical analysis of the proteomic output for one selected biological sample (thrombin‒activated platelets), performed in PEAKS 8.5, after de novo sequencing and PEAKS ID coupled with LFQ analysis. (A) The table containing the statistics for the total number (#) of MS and #MS/MS scans is complemented with the results for the filtration parameters, such as the peptide score, “A” score, protein score, #unique peptides/protein, and de novo ALC score, presented in Table 2. (B) The statistics for the filtered results are displayed in the table (panel A) which includes the FDR for PSM, peptides and proteins. The validation plot for “de novo” results corresponding to the residue local confidence is displayed in (a). The number (#) residues in the de novo sequences validated by confident database assignment are plotted as a function of the de novo residue local confidence (expressed as %) in (a). The validation plot for the residues found only by “de novo” sequences but not assigned by confident database search to any peptide assignment are shown in (b). (C) Venn diagram displaying the total number of proteins corresponding to the changes in the proteomic expression profiles in human platelets from one healthy donor under “resting” conditions (control) or ex‒vivo activated by thrombin corresponding to an FDR <1.2% for proteins and <0.2% for peptides, as calculated by the statistical LFDR built in algorithm in PEAKS 8.5. The complete list of proteins and details of mass spectrometry analysis for each of the proteomes of “resting “and “thrombin‒activated” platelets are provided in the Supplementary Table 1.
In Figure 4C the venn diagram displays the total number of proteins corresponding to the changes in the proteomic expression profiles in human platelets from one healthy donor under “resting” conditions (control) and ex‒vivo activation by thrombin. The representative example in Figure 4B&4C showed that the proteomic data successfully assigned 8302 peptides and 932 proteins having an FDR <1.2% for proteins and <0.2% for peptides, as calculated by the statistical LFDR built in algorithm in PEAKS 8.5. The complete list of proteins and details of mass spectrometry analysis for each of the proteomes of “resting “and “thrombin‒activated” platelets are provided in the Supplementary Table 1. The label‒free quantitative analysis of changes in the protein expression profiles from the “resting” and “thrombin‒activated” platelets was conducted first at the level of the precursor ion (MS1) using the LFQ quantitative module provided by PEAKS 8.5. As shown in Figure 5A, the intensity correlation plots displayed for different pairs of resting and thrombin‒activated platelets, after performing the LFQ analysis, are having the calculated Pearson’s correlation score (r=0.85) for the pair of “resting” and the “thrombin‒activated samples” which are expected to exhibit differences at the level of protein expression. In contrast, the two “alike” independent biological replicates corresponding either to the “thrombin‒activated” or to the “resting” platelets had the calculated correlation scores of 0.924 (in Figure 5B) and 0.939 (in Figure 5C) respectively, a proof of the high reproducibility of the developed proteomic assay. Additional parameters of mass spectrometric analysis are reported in Supplementary Table &2.

Figure 5 Label‒free quantitative analysis of changes in the protein expression profiles in “resting” and “thrombin‒activated” platelets.
The LFQ quantitative analysis used an independent “in silico” combined analysis (“Mud Pit” assay) to extract the combined total spectral counts (MS/MS) associated with the proteins from each biological sample. The LFQ analysis at the level of spectral counts retrieved the venn diagram displayed in Figure 5D, which shows a total of 924 proteins corresponding to the changes in the proteomic expression profiles in human platelets purified from three healthy donors. A total of 330 proteins are shared between the “resting” and “activated” platelets, while the 583 are having enhanced expression in the “activated” sample and only 11 proteins were mapped as being more expressed in the “resting” state. The data in the venn diagram was filtered to achieve an FDR<1.3% for proteins and <0.8% for peptides, as calculated by the Prophet peptide and protein algorithms built in algorithm in Scaffold (version 4.8.7, Proteome Software Inc.). The total 924 proteins characterized the proteome of “resting” and “thrombin‒activated” platelets represent about 17% from the total proteins reported to date for the platelets proteomes.50 The relative protein abundances identified in the whole platelet proteome was displayed as a clustered heat map after normalization of the corresponding weighted spectral counts (MS/MS level) using the built‒in algorithm from “per SPECtives” (version 2.0.6, Proteome Software Inc.) as shown in Figure 5E.
(A‒C) The intensity correlation plots displayed for different pairs of “resting” and “thrombin‒activated” platelets samples after performing the LFQ analysis for the selected proteomic data. The Pearson’s correlation score (r=0.85) indicates the expected proteomic differences between the “resting” and the “thrombin‒activated” samples” (in A) while the correlation scores of 0.924 (in B) and 0.939 (in C) indicate the high reproducibility of the proteomic changes monitored in biological triplicates for either the “resting” or the “thrombin‒activated” platelets. Additional parameters of mass spectrometric analysis are reported in Supplementary tables S1‒S2. (D) Venn diagram displaying the total number of proteins corresponding to the changes in the proteomic expression profiles in human platelets purified from three healthy donors for which an “in silico” combined analysis (“Mud Pit” assay) was performed to extract the combined total proteins from each biological sample. The data in the Venn diagram was filtered to achieve an FDR <1.3% for proteins and <0.8% for peptides, as calculated by the Prophet peptide and protein algorithms built in algorithm in Scaffold (version 4.8.7, Proteome Software Inc.). (E) The relative protein abundances identified in the whole platelet proteomes are displayed as a clustered heat map after normalization of the corresponding weighted spectral counts (MS/MS level) using the built‒in algorithm from “per SPECtives” (version 2.0.6, Proteome Software Inc.) as described in detail in the methods section. Only proteins which passed a selected significance statistical threshold (FDR <1.5 % for proteins, FDR<0.8% for peptide and p<0.05 by Fisher’s test followed by standard Benjamini‒Hochberg correction) were clustered using a single linkage method and a ranked‒based Euclidean distance‒metric in the representative heat map. The complete list of proteins and details of mass spectrometry analysis for each of the proteomes of “resting “and “thrombin‒activated” platelets are provided in the Supplementary Table &2. Only proteins which passed a selected significance statistical threshold (FDR <1.5 % for proteins, FDR<0.8% for peptide and p<0.05 by Fisher’s test followed by standard Benjamini‒Hochberg correction) were clustered using a single linkage method and a ranked‒based Euclidean distance‒metric in the representative heat map. The complete list of proteins and details of mass spectrometry analysis for each of the proteomes of “resting “and “thrombin‒activated” platelets are provided in the Supplementary Table &2. A zoomed view of one of the cluster of proteins having significant protein expression profiles (p<0.05, by Fisher exact test) is displayed in Figure 6.
Normalized weighted spectral counts quantified by label‒free analysis of protein expression profiles in the total proteomes from the “resting” and “thrombin‒activated” platelets. (C) Selected zoomed protein clusters views from the heat map displayed in Figure 5E highlight the major groups of proteins up‒regulated in the “thrombin‒activated” vs “resting” platelets, most of which were already reported by other studies on the proteomics of whole platelet, membrane‒derived proteins, microvesicles and releasates purified from platelets.1‒6 The relative protein abundances are displayed after normalization of the corresponding weighted spectral counts (MS/MS level) using the built‒in algorithm from “per SPECtives” (version 2.0.6, Proteome Software Inc.) as described in detail in the methods section. Details of the quantitative analysis for the selected proteomics data are presented in the Supplementary Table 2. Given the small volume of blood collected from each donor (8ml), the small sample size (three donors) and the small number of platelets used for each assay (2x108 platelets/sample), the 17% coverage of the major proteomic biomarkers characterizing the human platelets in resting and activated states represents an achievement in the proteomic profiling of human platelets. These data further support our developed proteomic assay, using one single‒step in solution digestion and peptides extraction coupled with nano LC/ESI/MS/MS on sensitive QExactive mass spectrometer, as a powerful tool for monitoring the qualitative and quantitative proteomic changes due to different pathophysiological conditions.
Development of an integrative bioinformatics and systems biology method to qualitatively and quantitatively assess the differential expression profile in the proteome of “thrombin‒activated” platelets
The changes in the proteomic landscapes were qualitatively and quantitatively analyzed using a combination of systems biology and bioinformatics approaches empowered by the ingenuity pathway analysis (IPA) and screening of the identified proteins and genes entities against the curated databases containing the platelets proteomes, including the “Adhesome”, “Exocarta”, “Reactome” and “Platelet Web”. About 50% of the our reproted total proteome of “thrombin‒activated” platelets was represented by up‒regulated and previously validated protein biomarkers involved in the pathways mediating the protease and thrombin activated receptor (PARs) and integrin signaling, the actin and rhoA linked to cytoskeleton signaling, and activated kinases pathways regulating the cellular motility, aggregation, procoagulation and degranulation.10‒45 The bioinformatics analysis for the whole proteome of “thrombin‒activated” platelets used first the “Reactome” database. Proteins IDs compiled in the Supplementary Table 1&2 were used to assign the gene ontology (GO) and the associated cellular pathways by employing the “Reactome” database (https://reactome.org/), as shown in Figures 7&8. The “Reactome” analysis retrieved the “hemostasis”, “platelets activation, signaling and aggregation” and “platelet degranulation” as the most significant pathways associated with the “thrombin‒activated” platelets (p<10‒10 for the gene entities, calculated by the statistics employed in the “Reactome” database) (Figure 7A). The proteome of “thrombin‒activated platelets” was thus exhibiting a significant increase in the platelet derived proteins involved in the maintenance of hemostasis, platelets activation, signaling, aggregation, degranulation and response to elevated platelet cytosolic Ca2+ pathways. In Figure 7B, Bioinformatics analysis for the whole proteome of “thrombin‒activated” platelets using the “Reactome” database. Proteins IDs compiled in the Supplementary Table 1&2 were used to assign the gene ontology (GO) and the associated cellular pathways using the “Reactome” database (https://reactome.org/). (A) The “Reactome” analysis retrieved the “hemostasis”, “platelets activation, signaling and aggregation” and “platelet degranulation” as the most significant pathways associated with the “thrombin‒activated” platelets (p<10‒10 for the gene entities, calculated by the statistics employed in the “Reactome” database). (B) The “platelets degranulation” pathway highlights in shaded yellow color at least 50% overlap between the experimentally identified proteins by the proteomic LFQ assay and the entities indexed in the “Reactome” database. False discovery rates (FDR) of less than 10‒8 were retrieved for the proteins clustered by the “Reactome” in the “platelet releasate secretory granules”, “platelet releasate cytosolic proteins”, “platelet dense granule membrane components”, “platelet alpha granules membrane‒derived and cytosolic proteins released by degranulation”. Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Table 3. The “platelets degranulation” pathway is displayed and the highlights in shaded yellow color are corresponding to the about 50% overlap between the experimentally identified proteins by the proteomic LFQ assay and the entities indexed in the “Reactome” database. False discovery rates (FDR) of less than 10‒8 were retrieved for the proteins clustered by the “Reactome” in the “platelet releasate secretory granules”, “platelet releasate cytosolic proteins”, “platelet dense granule membrane components”, “platelet alpha granules membrane‒derived and cytosolic proteins released by degranulation”. Proteins IDs having >95% probability assigned by the Prophet algorithm in Scaffold (shown in the Supplementary Table 2) were further used for gene ontology (GO) assignment and their associated cellular pathways by employing the “Reactome” database (https://reactome.org/).


Figure 7 Label‒free qualitative analysis of proteomes from “resting” and “thrombin‒activated” platelets.
The “Reactome” analysis generated an enriched view for “platelets activation, signaling and aggregation” and “platelet degranulation” pathways described in Figure 7. The statistical analysis highlighted the % overlap (in yellow color) between the experimental data and the proteins indexed in the “Reactome” as being part of the as the most significant signaling pathways associated with “platelets aggregation”, “GPVI‒mediated activated cascade”, “thrombin signaling through proteinase activated receptors (PAR)”, “response to elevated platelet cytosolic Ca2+) and “signaling amplification)” (p<0.05, using the “Reactome” algorithms). The statistically significant FDR scores characterizing the cellular pathways describing the activated platelets validated our new developed LFQ proteomic profiling as a robust assay for monitoring the proteomic landscape associated with platelets activation, signaling, degranulation and aggregation. Figure 8 Bioinformatics analysis for the whole proteome of “thrombin‒activated” platelets using the “Reactome” database. Proteins IDs compiled in the Supplementary Table 1&2 were used to assign the gene ontology (GO) and the associated cellular pathways using the “Reactome” database (https://reactome.org/). The “Reactome” analysis generated an enriched view for “platelets activation, signaling and aggregation” and “platelet degranulation” pathways described in Figure 7. The statistical analysis highlighted the % overlap (in yellow color) between the experimental data and the proteins indexed in the “Reactome” as being part of the as the most significant signaling pathways associated with “platelets aggregation”, “GPVI‒mediated activated cascade”, “thrombin signaling through proteinase activated receptors (PAR)”, “response to elevated platelet cytosolic Ca2+) and “signaling amplification) (p<0.05, using the “Reactome” algorithms). The statistically significant FDR scores validate the new developed LFQ proteomic profiling of “thrombin‒activated” platelets as a robust assay for monitoring the proteomic landscape associated with platelets activation, signaling, degranulation and aggregation. Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Table 3.
Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Tables 2‒4. One of the most important outcomes of the combined bioinformatics and systems biology approaches was reflected in the discovery of more than 50% overlap between the known and reported biomarkers of platelets degranulation and our experimental proteomic set describing the “thrombin‒activated” platelets. Proteins IDs having >95% probability assigned by the Prophet algorithm in Scaffold (shown in the Supplementary Table 2) were used for gene ontology (GO) assignment and their associated cellular pathways by employing the “Reactome”. Critical biomarkers like “von Willebrand factor”, “thrombospndin‒1”, SPARC, “coagulation factors V and XIII‒a”, “platelet glycoprotein V”, “platelet factor IV”, “fibrinogen‒alpha chain”, among many others shown in the Supplementary Tables 2‒4 and assigned by all platelet databases to be part of the degranulation of alpha‒granules pathway, were retrieved with very high confidence and low protein FDR (<1.0 %) by our new developed LFQ proteomics assay. Representative views of proteins known to be released from the alpha‒granules during platelets activation are displayed in Figure 9 Representative views of proteins known to be released from the alpha‒granules during platelets activation,1‒6 and their sequence coverage as reported by the Scaffold analysis (Proteome Software Inc.). In yellow are displayed the sequenced peptides while in green the amino acids with PTMs from each protein, used to calculate the sequence coverage. Additional proteins mapped to the alpha‒degranulation pathway are reported in the Supplementary Tables 2‒4.
Together with their sequence coverage as reported by the Scaffold analysis (Proteome Software Inc.). Additional proteins mapped to the alpha‒degranulation pathway are reported in the Supplementary Tables 2‒4. Complementary to the analysis employed by “Reactome” the proteome of “thrombin‒activated platelets” was screened for overlap with the proteins indexed by the “Exocarta” database (http://exocarta.org/) which complies the extended “exosomes and extracellular vesicles” derived platelets proteins. Bioinformatics analysis for the whole proteomes of “resting” and “thrombin‒activated” platelets by comparison with the reported proteomes derived from ectosomes, exosomes, microvesicles and other extracellular vesicles compiled in the “vesiclepedia” database is shown in the Figures 10‒13. This analysis showed that the proteome of “thrombin‒activated” platelets exhibits significant increase in the cellular compartments defining the platelet derived ectosomes, exosomes and microvesicles proteins (Figure 10A). Using the “Funrich” (functional enrichment) algorithm provided by the “exocarta” it was determined that about 58% of the total proteome of the “activated‒thrombin” platelets contain proteins derived from the exosomal vesicles (Figure 10A). Moreover, the “Funrich” analysis established that the “thrombin‒activated” platelet proteome was characterized by an increase expression of molecules involved in the signal transduction, cell communication, protein metabolism and energetic pathways, proportional with the involvement in the activation of platelets, cell signaling mediating the aggregation and degranulation (Figure 10B). Bioinformatics analysis for the whole proteomes of “resting” and “thrombin‒activated” platelets by comparison with the reported proteomes derived from ectosomes, exosomes, microvesicles and other extracellular vesicles compiled in the “vesiclepedia” database. Proteins IDs compiled in the Supplementary Table 1&2 were used to assign the gene ontology (GO) and the associated cellular locations and biological processes using the “Exocarta” database (http://exocarta.org/) containing the curated “vesiclepedia” proteomes. (A) About 58% of the total proteome of the “activated‒thrombin” platelets contain proteins derived from the exosomal vesicles. (B) The proteome of the “thrombin‒activated” platelets is characterized by increased expression of molecules involved in the signal transduction, cell communication, protein metabolism and energetic pathways, proportional with the involvement in the activation of platelets, cell signaling mediating the aggregation and degranulation. Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Table 3.
Details for the “Exocarta” bioinformatics analysis for the selected proteomics data are presented in the Supplementary Table 3. The venn diagram presented in Figure 11 Bioinformatics analysis for the whole proteomes of “resting” and “thrombin‒activated” platelets by comparison with the reported proteomes derived from ectosomes, exosomes, microvesicles and other extracellular vesicles compiled in the “vesiclepedia” database. Proteins IDs compiled in the Supplementary Tables 1&2 were used to assign the gene ontology (GO) and the associated cellular pathways using the “Exocarta” database (http://exocarta.org/) containing the curated “vesiclepedia” proteomes. The “Exocarta” analysis retrieved the overlapped proteomes with the “vesiclepedia” database by using selected parameters (shown to the right panel) for searching the presence of proteins derived from the microvesicles and exosomes released during platelet degranulation. False discovery rates (FDR) of less than 0.001 were retrieved for the proteins classified by the “Exocarta” database in the “platelets activation, signaling and aggregation”, “integrin‒mediated cellular signaling” and “thrombin‒activated protease (PAR) receptors pathways” (details in the Supplementary Table 3). Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Table 3. Further displays the enrichment of platelet derived ectosomes, exosomes, microvesicles proteins in the “thrombin‒activated platelets” proteomic data by comparison with the “vesiclepedia” proteomes provided by Exocarta database. The “Exocarta” analysis retrieved the overlapped proteomes with the “vesiclepedia” database by using the selected parameters (shown to the right panel, Figure 11). False discovery rates (FDR) of less than 0.001 were retrieved for the proteins classified by the “Exocarta” database in the “platelets activation, signaling and aggregation”, “integrin‒mediated cellular signaling” and “thrombin‒activated protease (PAR) receptors pathways” (see details in the Supplementary Table 3).

Figure 8 The proteome of “thrombin‒activated platelets” exhibits significant increase in the platelet derived proteins involved in the activation, signaling, aggregation and degranulation mediated by the GPVI and PAR receptors pathways.

Figure 9 The proteome of “thrombin‒activated platelets” exhibits significant increase expression in the proteins released from the platelets alpha‒granules, mediating the activation, signaling, and aggregation.
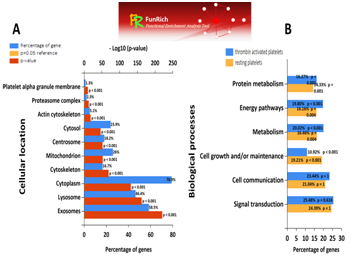
Figure 10 The proteome of “thrombin‒activated platelets” exhibits significant increase in the platelet derived ectosomes, exosomes and microvesicles proteins.

Figure 11 The proteome of “thrombin‒activated platelets” exhibits significant increase in the platelet derived ectosomes, exosomes, microvesicles proteins.
In addition to mapping the cellular location the “Exocarta” and the functional enrichment analysis performed with “Funrich” revealed that the proteome of “thrombin‒activated platelets” exhibited significant increase in the platelet activation, signaling, aggregation and degranulation pathways. Bioinformatics analysis for the whole proteomes of “thrombin‒activated” platelets (Figure 12). Bioinformatics analysis for the whole proteomes of “thrombin‒activated” platelets. Proteins IDs compiled in the Supplementary Table 1 & 2 were used to assign the gene ontology (GO) and the associated cellular pathways using the “Exocarta” database (http://exocarta.org/). The “Exocarta” analysis retrieved the “platelets activation, signaling and aggregation” and “platelet degranulation” as the most significant pathways associated with the “thrombin‒activated” platelets (p<0.001) for the gene entities, calculated by the statistics employed in the “Exocarta” database (left panel). An enriched protein‒protein interaction network displays the main gene‒protein players involved in the regulation of platelets activation, aggregation and degranulation (right panel). Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Table 3.

Figure 12 The proteome of “thrombin‒activated platelets” exhibits significant increase in the platelet activation, signaling, aggregation and degranulation pathways.
The “Exocarta” analysis retrieved the “platelets activation, signaling and aggregation” and “platelet degranulation” as the most significant pathways associated with the “thrombin‒activated” platelets (p<0.001) for the gene entities, calculated by the statistics employed in the “Exocarta” database left panel, Figure 12. These data further confirmed the analysis previously performed with the “Reactome” database. An enriched protein‒protein interaction network displays the main gene‒protein players involved in the regulation of platelets activation, aggregation and degranulation (right panel Figure 11). Moreover the “Exocarta” and “Funrich” analyses highlighted enrichment in “platelets activation, signaling and aggregation”, “thrombin/protease‒activated receptor (PAR)” PAR‒1 mediated thrombin signaling” and “integrins and proteoglycan‒mediated signaling” pathways (Figure 13A). A detailed analysis for the platelets‒derived proteins present in the proteomes of “resting” and “thrombin‒activated” platelets are displayed in the Scaffold analysis (from Proteome Software Inc.) in Figure 13B. Exocarta database and Scaffold software were used to conduct the bioinformatics analysis of the whole proteomes of “resting” and “thrombin‒activated” platelets. (A) Proteins IDs compiled in the Supplementary Table 1&2 were used to assign the gene ontology (GO) and the associated cellular pathways using the “Exocarta” database (http://exocarta.org/). The “Exocarta” analysis retrieved the “platelets activation, signaling and aggregation”, “thrombin/protease‒activated receptor (PAR)” PAR‒1 mediated thrombin signaling” and “integrins and proteoglycan‒mediated signaling” as leading pathways, having significant % of gene entities (>5%) involved in the thrombin‒mediated signaling in the activated platelets. (B) Detailed analysis for the platelets‒derived proteins identified in the proteomes of “resting” and “thrombin‒activated” platelets displayed by the Scaffold analysis (from Proteome Software Inc.). The final quantitative profiles associated with each sample are displayed as arrows, where red represents the down‒regulated and green is the up‒regulated trend calculated for each protein which passed the statistical significance threshold (p<0.05, by Fisher exact test). An enriched platelet derived glycoproteins view shows that the new developed nano LC/MS/MS assay retrieved 13 major platelets glycoproteins out of which three, i.e. PLF4, GP1BB and GP5, have significant increased fold change (>1.9, and p<0.05) in the “thrombin‒activated” vs the “resting” platelets. Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Table 2&3.

Figure 13 The proteome of “thrombin‒activated platelets” exhibits significant increase in the integrin‒and PAR1‒mediated signaling pathways and associated platelets glycoproteins and receptors.
The label‒free analysis data from three independent biological samples (control vs thrombin‒treated platelets) were exported from PEAKS 8.5 (Bioinformatics Solutions Inc.) into Scaffold where they were combined as one biological sample each using the “MudPit” built‒in option and were further re‒analyzed for the peptide and protein probabilities using the Prophet algorithm (see details in the methods section). The proteins having more than 95% probabilities were used to perform a label‒free analysis for the associated fold changes, using the normalized weighted spectral counts (MS/MS level). The Scaffold rescaled the protein ratios values using a log2 transformation, such that positive values reflected fold increases (in green) while the negative values reflected fold decreases (red). The final quantitative profile associated with each sample are displayed as arrows, where red represents the down‒regulated and green is the up‒regulated trend calculated for each protein which passed the statistical significance threshold (p<0.05, by Fisher exact test). An enriched platelet derived glycoproteins view shows that the new developed nano LC/MS/MS assay retrieved 13 major platelets glycoproteins out of the total 924 proteins having more than >95% identification probability, out of which three, i.e. PLF4, GP1BB and GP5, had significant increased fold change (>1.9, and p<0.05) in the “thrombin‒activated” vs the “resting” platelets. Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Table 2&3. We can argue with confidence that the bioinformatics analysis provided by the “Reactome” and the “Exocarta” together with the statistically significant FDR scores calculated for the “platelets activation, aggregation and degranulation” pathways, validate the our new developed LFQ platelets proteomic profiling as a robust assay for monitoring the proteomic landscape associated with platelets inter‒and intra‒cellular signaling during degranulation and aggregation.
A further in dept bioinformatics analysis of the cellular pathways and molecular functions in the proteome of “thrombin‒activated platelets” was performed using a quantitative method employing the ingenuity analysis pathway (IPA, Quigen), described by our lab elsewhere.59‒61 The samples were uploaded into the IPA application together with their rescaled log2 transformation of protein’s MS2 normalized weighted spectral counts ratios imported from Scaffold and per SPECtives software (Proteome Software Inc.). The probability of having a relationship between each IPA indexed biological function and the experimentally determined genes was calculated by a right‒tailed Fisher’s exact test. The level of significance was set to a “p‒value” of <0.05. Accordingly, the IPA analysis identified the molecular and cellular pathways from the IPA library of canonical pathways that were most significant to the dataset (Figure 14). Ingenuity Pathway Analysis (IPA) (Qiagen) predicted the up‒regulated (red) or downregulated (green) pathways in the “thrombin‒activated” vs “resting” platelets using the experimentally determined protein ratios, quantified using ≥ 1 peptides/protein, and their associated fold changes, after rescaling their values using a log2 transformation, such that positive values reflected fold increases while the negative values reflected fold decreases. The samples were uploaded into the IPA application together with their rescaled log2 transformation of protein’s MS2 normalized weighted spectral counts ratios imported from Scaffold and per SPECtives software (Proteome Software Inc.). The probability of having a relationship between each IPA indexed biological function and the experimentally determined genes was calculated by a right‒tailed Fisher’s exact test. The level of significance was set to a “p‒value” of <0.05. The graph shows the % overlap between the experimentally identified gene entities and the “knowledge” database provided by IPA displayed on x‒axis and the total number of gene entities assigned by IPA to each pathway on top of each bar histogram. Accordingly, the IPA analysis identified the molecular and cellular pathways from the IPA library of canonical pathways that were most significant to the dataset (‒log (p value)>2.0; in yellow on the graph). Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Table 4.
The IPA predicted up‒regulation (red) or down regulation (green) of each pathway in the “thrombin‒activated” vs “resting” platelets was based on the experimentally determined protein ratios, quantified using≥1 peptides/protein. The fold changes values for each protein were rescaled in IPA using a log2 transformation, such that positive values reflected fold increase while the negative values reflected fold decrease in the expression profiles. The proteome of “thrombin‒activated platelets” was predicted by IPA to exhibit significant increase in the integrin‒and PAR‒mediated signaling and changes in the cell motility mediated by cytoskeleton, RhoA and Rho‒GTP‒ase pathways (p<0.05, by Fisher exact test) (Figure 14 and Supplementary Table 4). In addition, IPA analysis further perform a functional enrichment for the proteomes and retrieved with high confidence cellular and molecular functions mediating the “aggregation of platelets” in the proteome of “thrombin‒activated” as compared with the “resting” platelets Figure 15. Thus, the quantitative analysis of protein expression profiles associated with a wide landscape of cellular functions was performed in IPA which assigned a “z‒score” function to all eligible canonical and cellular pathways. A “z<‒2 (in blue) represents significant down‒regulation while a z>2.0 (in orange) represents a significant up‒regulation of selected pathways. Cellular functions describing the “cell to cell signaling and interaction”, “immune cell trafficking”, “cellular movement” were mapped to be enriched in the proteome of the “thrombin‒activated” platelets (Figure 15A). An enriched protein network exhibiting the up‒regulation of “platelets aggregation” (z score of +5.1, in orange) was predicted by IPA to be the major network describing the cellular functions associated “cellular movement” and “cell to cell signaling” (Figure 15B and Supplementary Table 4). Detailed IPA analysis was extended to the main pathway which mediates the thrombin‒activated platelets aggregation pathway, i.e. the integrin‒and PAR1‒mediated signaling, predicted to be up‒regulated in the activated platelets (Figure 16A & Table 1). (A) The quantitative analysis of the protein expression profiles associated with a wide landscape of cellular functions was performed in IPA which assigned a “z‒score” function to all eligible canonical and cellular pathways. A “z<‒2 (in blue) represents significant down‒regulation while a z>2.0 (in orange) represents a significant up‒regulation of selected pathways. Cellular functions describing the “cell to cell signaling and interaction”, “immune cell trafficking”, “cellular movement” were mapped to describe the proteins in the proteome of the “thrombin‒activated” platelets. (B) An enriched protein network exhibiting the up‒regulation of “platelets aggregation” (z score of +5.1, in orange) was predicted by IPA to be the major network describing the cellular functions associated “cellular movement” and “cell to cell signaling”. Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Table 4. 52 proteins identified in the proteome of “thrombin‒activated” platelets are matched with the integrin signaling pathway by the knowledge database in IPA (Table 1). The IPA rescaled the protein ratios values using a log2 transformation, such that positive values reflected fold increases (shown as “Up or red color”) while the negative values reflected fold decreases (shown as “Down or green color”) as displayed in Table 1 and Figure 16A. The integrin signaling pathway has the assigned “activation z‒score” of “+5.28” (see details in Supplementary Table 4). An enhanced integrin glycoproteins view highlights that the new developed nano LC/MS/MS assay retrieved 6 major platelets integrins out of the total 924 identified proteins with >95% identification probability, out of which one, i.e. ITA2B, has significant increased fold change=2.4, p<0.05) in the “thrombin‒activated” vs the “resting” platelets. All the identified integrins were characterized by at least 2.4 increased fold change in the “thrombin‒activated” vs “resting” platelets. (Figure 16B, Table 1 and Supplementary Tables 2 &4). (A) IPA predicted the up‒regulation of “integrin‒mediated signaling”, which is a major canonical path way regulated in the thrombin‒activated platelets. The canonical pathway is displayed by IPA using the experimentally determined protein ratios, quantified using ≥1 peptides/protein, and their associated fold changes, shown as “Experiment Log (protein ratio”). The IPA rescaled the protein ratios values using a log2 transformation, such that positive values reflected fold increases (in red) while the negative values reflected fold decreases (green). The integrin signaling pathway has IPA assigned “activation z‒scores”>2.0. (B) A zoomed integrin glycoproteins view displayed by Scaffold analysis shows that the new developed nanoLC/MS/MS assay retrieved 6 major platelets glycoproteins out of the total 924 identified proteins with >95% identification probability, out of which one, i.e. ITA2B, has significant increased fold change=2.4, p<0.05) in the “thrombin‒activated” vs the “resting” platelets. All the identified integrins were characterized by at least 2.4 increase fold change in the “thrombin‒activated” vs “resting” platelets. The proteins involved in the integrin signaling pathways and their associated fold changes are displayed in the Table 1. Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Supplementary Tables 2 &4.
Symbol |
Entrez gene name |
Expr log ratio |
Expected |
Location |
Type(s) |
RAP1A |
RAP1A, member of RAS oncogene family |
25.632 |
Up |
Cytoplasm |
enzyme |
ARF5 |
ADP ribosylation factor 5 |
24.019 |
Down |
Cytoplasm |
enzyme |
CAPNS1 |
calpain small subunit 1 |
24.019 |
Up |
Cytoplasm |
peptidase |
ARF4 |
ADP ribosylation factor 4 |
23.391 |
Down |
Cytoplasm |
enzyme |
CRKL |
CRK like proto-oncogene, adaptor protein |
22.932 |
Up |
Cytoplasm |
kinase |
GRB2 |
growth factor receptor bound protein 2 |
22.932 |
Up |
Cytoplasm |
kinase |
MAPK1 |
mitogen-activated protein kinase 1 |
21.932 |
Up |
Cytoplasm |
kinase |
RAP2B |
RAP2B, member of RAS oncogene family |
20.932 |
Up |
Plasma Membrane |
enzyme |
WAS |
Wiskott-Aldrich syndrome |
20.932 |
Up |
Cytoplasm |
other |
RALB |
RAS like proto-oncogene B |
19.932 |
Up |
Cytoplasm |
enzyme |
RRAS |
RAS related |
19.932 |
Up |
Cytoplasm |
enzyme |
ACTG2 |
actin, gamma 2, smooth muscle, enteric |
4.605 |
Up |
Cytoplasm |
other |
ARF3 |
ADP ribosylation factor 3 |
4.392 |
Down |
Cytoplasm |
enzyme |
ACTA1 |
actin, alpha 1, skeletal muscle |
3.985 |
Up |
Cytoplasm |
other |
MYL12A |
myosin light chain 12A |
3.222 |
Up |
Cytoplasm |
other |
MYL12B |
myosin light chain 12B |
3.196 |
Up |
Cytoplasm |
other |
VASP |
vasodilator stimulated phosphoprotein |
2.615 |
Down |
Plasma Membrane |
other |
ACTN2 |
actinin alpha 2 |
2.382 |
Up |
Nucleus |
transcription regulator |
ACTB |
actin beta |
2.343 |
Up |
Cytoplasm |
other |
ACTG1 |
actin gamma 1 |
2.203 |
Up |
Cytoplasm |
other |
ARF1 |
ADP ribosylation factor 1 |
2.138 |
Down |
Cytoplasm |
enzyme |
MYL9 |
myosin light chain 9 |
2.115 |
Up |
Cytoplasm |
other |
ARPC5 |
actin related protein 2/3 complex subunit 5 |
1.874 |
Up |
Cytoplasm |
other |
MYLK |
myosin light chain kinase |
1.585 |
Up |
Cytoplasm |
kinase |
RAP1B |
RAP1B, member of RAS oncogene family |
1.585 |
Up |
Cytoplasm |
enzyme |
ARPC1B |
actin related protein 2/3 complex subunit 1B |
1.495 |
Up |
Cytoplasm |
other |
LIMS1 |
LIM zinc finger domain containing 1 |
1.471 |
Up |
Plasma Membrane |
other |
ACTN3 |
actinin alpha 3 (gene/pseudogene) |
1.354 |
Up |
Plasma Membrane |
other |
SRC |
SRC proto-oncogene, non-receptor tyrosine kinase |
1.354 |
Up |
Cytoplasm |
kinase |
ACTN4 |
actinin alpha 4 |
1.331 |
Up |
Cytoplasm |
transcription regulator |
ZYX |
zyxin |
1.31 |
‒ |
Plasma Membrane |
other |
ARPC2 |
actin related protein 2/3 complex subunit 2 |
1.237 |
Up |
Cytoplasm |
other |
GSN |
gelsolin |
1.184 |
‒ |
Extracellular Space |
other |
ARPC4 |
actin related protein 2/3 complex subunit 4 |
1.087 |
Up |
Cytoplasm |
other |
ACTN1 |
actinin alpha 1 |
1.07 |
Up |
Cytoplasm |
transcription regulator |
CAPN1 |
calpain 1 |
1.059 |
Up |
Cytoplasm |
peptidase |
RAC2 |
Rac family small GTPase 2 |
0.848 |
Up |
Cytoplasm |
enzyme |
ACTR3 |
ARP3 actin related protein 3 homolog |
0.817 |
Up |
Plasma Membrane |
other |
ACTR2 |
ARP2 actin related protein 2 homolog |
0.807 |
Up |
Plasma Membrane |
other |
VCL |
vinculin |
0.773 |
Up |
Plasma Membrane |
enzyme |
ILK |
integrin linked kinase |
0.755 |
Up |
Plasma Membrane |
kinase |
PFN1 |
profilin 1 |
0.696 |
Up |
Cytoplasm |
other |
PARVB |
parvin beta |
0.596 |
Down |
Cytoplasm |
other |
ARPC3 |
actin related protein 2/3 complex subunit 3 |
0.585 |
Up |
Cytoplasm |
other |
ITGB1 |
integrin subunit beta 1 |
0.585 |
Up |
Plasma Membrane |
transmembrane receptor |
TLN1 |
talin 1 |
0.488 |
Up |
Plasma Membrane |
other |
ITGA2B |
integrin subunit alpha 2b |
0.466 |
Up |
Plasma Membrane |
transmembrane receptor |
ITGB3 |
integrin subunit beta 3 |
0.464 |
Up |
Plasma Membrane |
transmembrane receptor |
ITGA6 |
integrin subunit alpha 6 |
0.263 |
Up |
Plasma Membrane |
transmembrane receptor |
RHOA |
ras homolog family member A |
0.222 |
Up |
Cytoplasm |
enzyme |
CDC42 |
cell division cycle 42 |
0.152 |
Up |
Cytoplasm |
enzyme |
CTTN |
cortactin |
0 |
Up |
Plasma Membrane |
other |
Table 1 Ingenuity pathway analysis (ipa) predicted up-regulation of “integrin-mediated signaling”, as a major canonical pathway regulated by thrombin activation in the platelets proteome

Figure 14 The proteome of “thrombin‒activated platelets” exhibits significant increase in the integrin‒and PAR1‒mediated signaling and changes in the cell motility mediated by cytoskeleton, RhoA and Rho‒GTP‒ase pathways.

Figure 15 The proteome of “thrombin‒activated platelets” is enriched in proteins network associated with cellular and molecular functions mediating the “aggregation of platelets”.

Figure 16 The proteome of “thrombin‒activated platelets” exhibits significant increase in the integrin‒and PAR1‒mediated signaling.
A further bioinformatics analysis revealed that the proteome of “thrombin‒activated platelets” exhibits significant increase in the tyrosine and serine/threonine kinase mediated cellular signaling (Figures 17). Detailed analysis for the kinase protein family present in the proteomes of “resting” and “thrombin‒activated” platelets were performed in Scaffold (from Proteome Software Inc.). The proteins having more than 95% identification probability were used to perform a label‒free analysis for the associated fold changes, using the normalized weighted spectral counts (MS/MS level), as described previously for other proteins displayed in Figure 13. A zoomed view displaying 11 out of 42 kinases identified in our proteomic set is displayed in Figure 17, Detailed analysis for the kinase protein family present in the proteomes of “resting” and “thrombin‒activated” platelets are displayed in the Scaffold analysis (from Proteome Software Inc.). The proteins having more than 95% identification probability were used to perform a label‒free analysis for the associated fold changes, using the normalized weighted spectral counts (MS/MS level). The final quantitative profile associated with each sample are displayed as arrows, where red represents the down‒regulated and green is the up‒regulated trend calculated for each protein which passed the statistical significance threshold (p<0.05, by Fisher exact test). An enhanced view displaying 11 out of 42 identified kinases shows that most of the identified protein kinases exhibited more than two‒fold increase in their expression profiles in the “thrombin‒activated” vs “resting” platelets. The Scaffold rescaled the protein ratios values using a log2 transformation, such that positive values reflected fold increases (in green) while the negative values reflected fold decreases (red). Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Table 2 and methods section. showing that these proteins had more than two‒fold increase in their expression profiles in the “thrombin‒activated” vs “resting” platelets. We recovered a high overlap of proteins members with the kinome family reported already by other proteomic analyses,20‒30 that are also indexed in many other research studies related to the proteomic profiling of agonists activated platelets, such as the “Adhesome” and “Platelet Web” databases, as described later in the results section. Other major pathways predicted by IPA to be up‒regulated in the proteome of “thrombin‒activated platelets” were mapped to the cytoskeleton, actin and RhoA mediated cellular signaling (Figure 18). Ingenuity Pathway Analysis (IPA) predicted the up‒regulation of “actin and cytoskeleton mediated signaling” in (A) and “RhoA mediated cytoskeleton signaling and cell motility” (in B), which are major canonical pathways regulated by thrombin activation in platelets. The canonical pathways are displayed by IPA using the experimentally determined protein ratios, quantified using ≥1 peptides/protein, and their associated fold changes. There is at least 21% overlap between the experimentally identified gene entities in the whole proteome of the platelets and the knowledge‒based IPA database. The IPA rescaled the protein ratios values (derived from MS1 or MS2 LFQ analysis) using a log2 transformation, such that positive values reflected fold increases (in red) while the negative values reflected fold decreases (green) (see methods for details). Both pathways have IPA assigned “activation z‒scores” >2.0, as shown in the Table 2 and Supplementary Table 4. 47proteins identified in the proteome of “thrombin‒activated” platelets are matched with the actin cytoskeleton signaling pathway by the knowledge database in IPA, as shown in Table 2. The cytoskeleton signaling pathway the assigned an “activation z‒score” of “+5.94” (see details in Supplementary Table 4). An independent systems biology approach aimed to validate the molecular functions and the cellular pathways in the “thrombin‒activated” vs “resting” platelets employed the Platelet Web” (http://plateletweb.bioapps.biozentrum.uni‒wuerzburg.de/plateletweb.php) database, shown in Figure 19. “PlateletWeb” (http://plateletweb.bioapps.biozentrum.uni‒wuerzburg.de/plateletweb.php) database was used to conduct the bioinformatics analysis of the whole proteome from the “thrombin‒activated” platelets. More than 70% of the proteins identified in the “thrombin‒activated platelets” are overlapped with signaling networks comprised of kinases, proteases, phosphatases, integrins and platelets derived glycoproteins, curated in the system biology networks at the Platelet Web database. Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Table 4.
Symbol |
Entrez gene name |
Expr log ratio |
Expected |
Location |
Type(s) |
MYH10 |
myosin heavy chain 10 |
25.253 |
Up |
Cytoplasm |
enzyme |
EZR |
ezrin |
23.101 |
Up |
Plasma Membrane |
other |
CRKL |
CRK like proto-oncogene, adaptor protein |
22.932 |
Up |
Cytoplasm |
kinase |
GRB2 |
growth factor receptor bound protein 2 |
22.932 |
Up |
Cytoplasm |
kinase |
MAPK1 |
mitogen-activated protein kinase 1 |
21.932 |
Up |
Cytoplasm |
kinase |
WAS |
Wiskott-Aldrich syndrome |
20.932 |
Up |
Cytoplasm |
other |
KNG1 |
kininogen 1 |
19.932 |
‒ |
Extracellular Space |
other |
RRAS |
RAS related |
19.932 |
Up |
Cytoplasm |
enzyme |
ACTG2 |
actin, gamma 2, smooth muscle, enteric |
4.605 |
Up |
Cytoplasm |
other |
ACTA1 |
actin, alpha 1, skeletal muscle |
3.985 |
Up |
Cytoplasm |
other |
MYL12A |
myosin light chain 12A |
3.222 |
Up |
Cytoplasm |
other |
MYL12B |
myosin light chain 12B |
3.196 |
Up |
Cytoplasm |
other |
CSK |
C-terminal Src kinase |
2.585 |
Up |
Cytoplasm |
kinase |
ACTN2 |
actinin alpha 2 |
2.382 |
Up |
Nucleus |
transcription regulator |
ACTB |
actin beta |
2.343 |
Up |
Cytoplasm |
other |
ACTG1 |
actin gamma 1 |
2.203 |
Up |
Cytoplasm |
other |
MYL6 |
myosin light chain 6 |
2.198 |
Up |
Cytoplasm |
enzyme |
MYL9 |
myosin light chain 9 |
2.115 |
Up |
Cytoplasm |
other |
ARPC5 |
actin related protein 2/3 complex subunit 5 |
1.874 |
Up |
Cytoplasm |
other |
DIAPH1 |
diaphanous related formin 1 |
1.837 |
Up |
Plasma Membrane |
other |
MSN |
moesin |
1.784 |
Up |
Plasma Membrane |
other |
MYLK |
myosin light chain kinase |
1.585 |
‒ |
Cytoplasm |
kinase |
ARPC1B |
actin related protein 2/3 complex subunit 1B |
1.495 |
Up |
Cytoplasm |
other |
ACTN3 |
actinin alpha 3 (gene/pseudogene) |
1.354 |
Up |
Plasma Membrane |
other |
ACTN4 |
actinin alpha 4 |
1.331 |
Up |
Cytoplasm |
transcription regulator |
ARPC2 |
actin related protein 2/3 complex subunit 2 |
1.237 |
Up |
Cytoplasm |
other |
GSN |
gelsolin |
1.184 |
Down |
Extracellular Space |
other |
ARPC4 |
actin related protein 2/3 complex subunit 4 |
1.087 |
Up |
Cytoplasm |
other |
ACTN1 |
actinin alpha 1 |
1.07 |
Up |
Cytoplasm |
transcription regulator |
FN1 |
fibronectin 1 |
1 |
Up |
Extracellular Space |
enzyme |
CFL1 |
cofilin 1 |
0.883 |
Down |
Nucleus |
other |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase type 2 alpha |
0.848 |
Up |
Cytoplasm |
kinase |
RAC2 |
Rac family small GTPase 2 |
0.848 |
Up |
Cytoplasm |
enzyme |
ACTR3 |
ARP3 actin related protein 3 homolog |
0.817 |
Up |
Plasma Membrane |
other |
ACTR2 |
ARP2 actin related protein 2 homolog |
0.807 |
Up |
Plasma Membrane |
other |
VCL |
vinculin |
0.773 |
Up |
Plasma Membrane |
enzyme |
ROCK2 |
Rho associated coiled-coil containing protein kinase 2 |
0.737 |
Up |
Cytoplasm |
kinase |
MYH9 |
myosin heavy chain 9 |
0.721 |
Up |
Cytoplasm |
enzyme |
PFN1 |
profilin 1 |
0.696 |
Up |
Cytoplasm |
other |
ARPC3 |
actin related protein 2/3 complex subunit 3 |
0.585 |
Up |
Cytoplasm |
other |
ITGB1 |
integrin subunit beta 1 |
0.585 |
Up |
Plasma Membrane |
transmembrane receptor |
TLN1 |
talin 1 |
0.488 |
Up |
Plasma Membrane |
other |
FLNA |
filamin A |
0.35 |
Up |
Cytoplasm |
other |
RHOA |
ras homolog family member A |
0.222 |
Up |
Cytoplasm |
enzyme |
CDC42 |
cell division cycle 42 |
0.152 |
Up |
Cytoplasm |
enzyme |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
0 |
Down |
Cytoplasm |
other |
CYFIP1 |
cytoplasmic FMR1 interacting protein 1 |
-0.415 |
‒ |
Cytoplasm |
other |
Table 2 Ingenuity pathway analysis (ipa) predicted up-regulation of “actin cytoskeleton signaling”, as a major canonical pathway regulated by thrombin activation in the platelets proteome

Figure 17 The proteome of “thrombin‒activated platelets” exhibits significant increase in the kinase mediated cellular signaling.

Figure 18 The proteome of “thrombin‒activated platelets” exhibits significant increase in the cytoskeleton, actin and RhoA mediated cellular signaling.

Figure 19 The proteome of “thrombin‒activated platelets” exhibits significant increase in the number of kinases, phosphatases, proteases and adhesion receptors mediating the cellular signaling.
It was found that more than 70% of the proteins identified in the “thrombin‒activated platelets” are overlapped with signaling networks comprised of kinases, proteases, phosphatases, integrins, adhesion receptors mediating the cellular signaling and platelets derived glycoproteins curated in the system biology networks at the Platelet Web database (see details in the Supplementary Table 4 where an independent network related to the integrin ITA2B is also displayed). In addition, the Platelet Web systems biology approach highlighted the proteome of “thrombin‒activated platelets” exhibits significant increase in the number of “cluster of differentiation” (CD) adhesion receptors mediating the cellular adhesion and platelets aggregation (Figure 20 and Supplementary Table 2‒4). “Platelet Web” (http://plateletweb.bioapps.biozentrum.uni‒wuerzburg.de/plateletweb.php) database was used to conduct the bioinformatics analysis of the whole proteome from the “thrombin‒activated” platelets. (A‒C) The “Platelet Web” systems biology retrieved the molecular interactions and the networks describing the protein‒protein interactions regulating the CD9, CD226 and CD36 interactions, reportedly to mediate the platelets adhesion and aggregation when activated by thrombin.5‒10 (D) Detailed analysis for the platelets‒derived “CD” proteins present in the proteomes of “resting” and “thrombin‒activated” platelets displayed in the Scaffold analysis (from Proteome Software Inc.). The final quantitative profiles associated with each sample are displayed as arrows, where red represents the down‒regulated and green is the up‒regulated trend calculated for each protein which passed the statistical significance threshold (p<0.05, by Fisher exact test). Details of the bioinformatics analysis for the selected proteomics data are presented in the Supplementary Tables 2‒4. The “Platelet Web” systems biology retrieved the molecular interactions and the networks describing the protein‒protein interactions regulating the CD9, CD226 and CD36 interactions, reportedly to mediate the platelets adhesion and aggregation when activated by thrombin.5‒10

Figure 20 The proteome of “thrombin‒activated platelets” exhibits significant increase in the number of “cluster of differentiation” (CD) adhesion receptors mediating the cellular adhesion and paltelets aggregation.
As have been already proved by the other bioinformatics approaches described above, the high % of overlap between the experimentally identified proteins and the platelets signaling networks provided by the Platelet Web, complemented by their functional annotations in the “platelets activation and signaling” pathways validate the new developed LFQ platelets proteomic profiling as a robust assay for monitoring the proteomic landscape associated with the activation of inter‒and intra‒cellular signaling pathways during platelets degranulation and aggregation. A final bioinformatics approach used the “Adhesome” database which is a curated library of proteins and receptors involved in cell‒cell communication and adhesion, signaling and protein‒protein interactions involving integrins and many kinase family members (http://www.adhesome.org/). Proteins IDs compiled in the Supplementary Table 1&2 were used to assign the gene ontology (GO) and the associated cellular pathways using the “Adhesome” database. The “Funrich” functional enrichment program provided by “Exocarta” was used to generate the overlap between the biological pathways in the “adhesome” and the proteins identified in the “thrombin‒activated” platelets. A statistical significance test was applied to assess the significance of the overlap between the experimental proteomic data set and the proteins indexed in the “Adhesome”, where a p‒value of <0.001 was categorized to be significant for the overlap. The “Adhesome” and “Funrich” analyses showed that the proteome of “thrombin‒activated platelets” exhibits significant increase in the integrin, proteoglycan/syndecan, plasma membrane receptors, VEGFR mediated signaling and glycoproteins and receptors associated with the cell signaling pathways (Figure 21A). As shown in Figure 21B, the 51 proteins from the experimental proteomic dataset overlapped with the proteome indexed in the “Adhesome”, which correspond to about 22% from the reported adhesion receptors and adaptors proteins, intrinsic proteins and kinases involved in “integrin cell surface interactions”, “TRAIL mediated signaling” and “integrins and proteoglycan‒mediated signaling” pathways. The gene symbols describing the 51 protein entities are functional annotated and presented in the Supplementary Table 4. “Adhesome” database was used to conduct the bioinformatics analysis of the whole proteome “thrombin‒activated” platelets. (A) Proteins IDs compiled in the Supplementary Tables 1&2 were used to assign the gene ontology (GO) and the associated cellular pathways using the “Adhesome” database (http://www.adhesome.org/). The “Funrich” functional enrichment program provided by “Exocarta” was used to generate the overlap between the biological pathways in the “adhesome” the proteins identified in the “thrombin‒activated” platelets. A statistical significance test was applied was applied to assess the significance of the overlap between the experimental proteomic data set and the proteins indexed in the “Adhesome”, where a p‒value of <0.001 was categorized to be significant for the overlap. (B) 51 proteins from the experimental proteomic dataset overlapped with the proteome indexed in the “Adhesome”, which correspond to about 22% from the reported adhesion receptors and adaptors proteins, intrinsic proteins and kinases involved in “integrin cell surface interactions”, “TRAIL mediated signaling” and “integrins and proteoglycan‒mediated signaling” pathways. The gene symbols describing the 51 protein entities are functional annotated and presented in the Supplementary Table 4.
The research presented herein employed a simple nano LC‒ESI/MS/MS sequencing of peptides derived from the “in solution,” one step digestion of the whole proteomes of “resting” and “thrombin‒activated” platelets, extracted by a single‒one step solubilization procedure using denaturing reagents. We successfully employed a method for platelets lysis in absence of any detergent, i.e. a cryogenic lysis in liquid nitrogen, followed by protein solubilization under denaturant conditions (in 50mM ammonium bicarbonate buffer, pH 8.5 with 8Murea) that enabled the solubilization of key membrane‒bound proteins, in addition to the cytosolic and organelles‒derived proteins secreted in the microparticles and found in the platelets releasates during thrombin activation. The new, developed assay employed the nano LC‒ESI/MS/MS sequencing of peptides derived from the “in solution,” one step digestion of the whole platelets proteomes by means of three enzymes (tryptic/Lys‒C/Glu‒C), and a Q‒Exactive quadrupole orbitrap mass spectrometer coupled with the label free quantification (LFQ) analysis. The changes in the proteomic landscapes were qualitatively and quantitatively analyzed using a combination of systems biology and bioinformatics approaches empowered by the ingenuity pathway analysis (IPA) and screening of the identified proteins and genes entities against the curated databases containing the platelets proteomes, including the “Adhesome”, “Exocarta,” “Reactome,” and “Platelet Web”. We were able to cover more than 17% (corresponding to 924 total proteins), from the up‒to date reported proteome of human platelets under resting or agonist‒activated conditions. The changes in the proteomic landscapes were qualitatively and quantitatively analyzed using a combination of systems biology and bioinformatics approaches empowered by the ingenuity pathway analysis (IPA) and screening of the identified proteins and genes entities against the curated databases containing the platelets proteomes, including the “Adhesome,” “Exocarta,” “Reactome,” and “Platelet Web.” The high, >50% overlap between the experimentally identified proteins and the platelets signaling networks provided by the databases indexing the platelets proteomes validated the new developed LFQ proteomic profiling as a robust assay for monitoring the proteomic landscape associated with platelets inter‒and intra‒cellular signaling during degranulation and aggregation. The outcome of the employed systems biology approaches further advocates for the implementation of our LFQ proteomics assay in clinical labs focused on understanding platelets’ response to thrombin activation under pathophysiological circumstances, especially when potent thrombin inhibitors and novel therapies are investigated for the treatment of inflammatory thrombosis, heart attack, stroke, and other complications of ACS syndrome heavily or partly mediated by platelets aggregation.
Support for this project was provided by the CUNY Research Scholars Program, which is funded by The City University of New York and the New York City Mayor's Office.
The authors declared that there is no conflict of Interest.

©2018 Gonzalez, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.