MOJ
eISSN: 2374-6920


Research Article Volume 2 Issue 4
1Department of Biology, University of Isfahan, R Iran
2Department of Pharmaceutical Biotechnology, Shiraz University of Medical Sciences, IR Iran
Correspondence: Sadeq Vallian, Professor of Human Molecular Genetics Division of genetics, Department of Biology, Faculty of Science, University of Isfahan, Isfahan, Iran, Tel +983117932456
Received: March 07, 2015 | Published: October 19, 2015
Citation: Mehraban MH, Ghasemi Y, Vallian S. A computational comparative study of α -glucosidase enzyme divergence. MOJ Proteomics Bioinform. 2015;2(4):139-141. DOI: 10.15406/mojpb.2015.02.00057
α-glucosidases (α-Gls) catalyze the last step of carbohydrate digestion in mammals and release glucose in bloodstream, which results in the increased blood glucose level. The evolution and classification of this enzyme has been a matter of debate. In the present study the amino acid sequences of α-Gls from 12 species were aliened and the Phylogenetic trees were constructed. The data indicated that only the mammalian enzyme contained the glucoamylase region. Five amino acid regions were found to be the conservative blocks of mammalian α-Gls. These blocks were not fully conserved in plants, fungi and bacteria. The results were in favor of Chiba`s classification despite the Ile→Thr substitution in Aspergillus Niger and Ala→Pro substitution in chimpanzee and human α-Gls. The chimpanzee α-Gls showed the most similarity to human enzyme. Plants, fungi, bacteria, and the mammalian α-Gls seemed to separately create a specific class. Together, the data suggest that the eukaryotic enzyme had been diverged significantly from the prokaryotic α-Gls since the separation from the common ancestor.
Keywords: α –glucosidase, phylogenetic trees, evolution, multiple alignments
α-Glucosidase (α-Gls) enzymes have a crucial role in digestion of carbohydrates and biosynthesis of glycoprotein.1,2 Theses enzymes hydrolyze terminal glycoside bonds and release α-glucose from the substrate chain. They are found in prokaryotic cells, plants and mammalian tissues such as liver, blood and especially intestine.1 The last step of carbohydrate digestion is catalyzed by α-glucosidases (namely maltase-glucoamylase or MGAM in mammals) which results in liberation of glucose. In Escherichia coli and viruses such as human immunodeficiency virus (HIV) they are necessary for cell wall and envelope construction.2 In plants, the glucose produced by the activity of these enzymes is used as an energy source for cellular growth and development. These enzymes are distributed in plant tissues like seeds, fruits, leaves and roots. In the absence of α-amylase it is suggested that α-Gls can initiate the degradation of natural starch granules in barely seeds and pea chloroplasts.3-5 Their important role in treating degenerative diseases such as diabetes mellitus type-II and HIV has been proved in many studies.6-11 Although, the enzymes exist in monomeric form in prokaryotes, plants and primary eukaryotes, hetero and homodimeric structures were found in advanced Eukaryotes.1,12 Therefore, the classification of α-Gls enzyme is of great importance for enzymologists and evolutionists. According to Chiba`s classification, Escherichia coli and other bacterial species constitute family I of α-Gls.13 On the other hand, α-Gls from mammalian tissues, plants, Candida tsukubaensis, and Mucor javanicus constitute the family II. The basis of this classification is the amino acid conservative blocks existed in these enzymes. The amino acid block of DLVINH, EVAH and YIENHD seems to be present in class I. However the conservative blocks of class II are GIWADMNEV and GADICGF.1 In the present study the aim was to assess the similarities between the two classes using evolutionarily relationship and Phylogenetic data. Currently it is not known how α-Gls had been evolved and how many species have the enzyme, and what the amino acid sequence of the ancestor was. Answer to these questions will facilitate identification of primitive and specialized aspects of α-Gls structure, function and regulation.
Amino acid sequences of E. coli, Sulfolobus solfataricus, yeast (Candida tsukubaensis), barley (Hordeum vulgare), Aspergillus Niger, spinach (Spinacia oleracea), sugar beet (Beta vulgaris), mouse (Mus musculus), rat (Rattsu norvegicus), chimpanzee (Pan Troglodyte) and human (Homo sapiens) α-glucosidase enzyme were retrieved from Uniprot (protein data base). The full sequence of each entity was used. The sequences were aligned and the Phylogenetic trees were constructed. Clustal X version 2.114 was used for multiple alignment and Phylogenetic trees were constructed using Mega 6 software.15 Trees were created using maximum-likelihood approach, and 1000 bootstrap replicates were conducted.
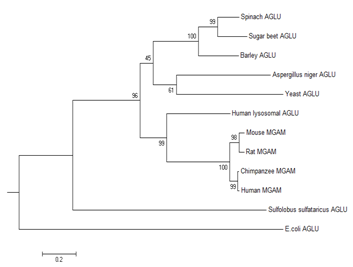
In the present study a comparative investigation on the divergence of α-glucosidase (α-Gls) enzyme was performed. The complete amino acid sequences of different species as stated in the materials and methods were aligned. The mammalian α-Gls enzymes are heterodimeric proteins with α-glucosidic activity. The enzymes contain a region responsible for the catalytic site for an enzyme termed glucoamylase. The comparative results from the present investigation showed that human and chimpanzee α-glucosidases had the closest similarity, and may have come from the same ancestor. There are two conserve P-type domains in human MGAM located in positions 88-134 and 954-1000,16 which is completely conserved in chimpanzee`s enzyme. These domains have some synonymous amino acid alterations in rat and mouse MGAM. Interestingly, these domains are absent in plants and bacterial counterparts. There are two catalytic aspartic acid residues in mammalian enzymes which due to their nucleophilic nature has a major role in hydrolyzing glucosidic chains.16 One of them which are located in position 529 is conserved among all the species studied. This residue is in the maltase region of mammalian MGAM and is conserved in plants and bacterial enzymes, which improves the importance of this residue in catalytic site. The other aspartic acid residue is in position 1420 which is located in glucoamylase region of mammalian MGAM. This is a remarkable feature of mammalian enzymes which is absent in plants and bacterial α-Gls. These characteristics make mammalian α-Gls a unique class which has different structure and function in comparison to plants and bacterial enzymes. Rat and mouse α-Gls showed a significant similarity with human enzyme, but obviously they had not been diverged from a common ancestor. It was noteworthy that five amino acid regions were found to be the conservative blocks of mammalian α-Gls which was summarized in (Table 1). These blocks were not fully conserved in plants, fungi and bacteria (Figure 1). Aspergillus Niger, spinach, sugar beet, barley and yeast (Candida tsukubaensis) α-Gls exist in monomeric form. The plant enzyme proved to create a different subclass since sugar beet; barley and spinach shared more similar amino acid sequences with each other. On the other hand, Aspergillus Niger and yeast may be considered as the other subclass due to their sequence homology. According to Chiba`s classification, plants, mammalian, yeast and Aspergillus Niger α-Gls are considered the family II of α-Gls as they all have a conservative block of GADICGF.1,13
Our results were in favor of the above theory despite the Ile→Thr substitution in Aspergillus Niger and Ala→Pro substitution in chimpanzee and human α-Gls. However, due to the presence of glucoamylase site in mammalian enzymes, the classification could be changed in a way that the mammalian enzymes would be considered as an independent class. The results showed that E. coli and Sulfolobus sulfataricus α-Gls sequences were significantly different from plants and mammalian α-Gls Interestingly it was proved that despite the mammalian α-Gls other enzyme counterparts do not contain the conservative blocks which was part of glucoamylase region of mammalian α-Gls. These conservative blocks were summarized in (Table 2). It might be concluded that during the evolutionary state of mammals, this region was inserted or had some de novo mutations which resulted in a perfect active glucoamylase that could produce β-glucose. The classification of α-Gls enzyme is of great importance to understand the structure and function of this crucial enzyme, and might help evolutionist understand the origin of this enzyme. As mentioned above, the inhibition of this enzyme had a major role in treating diabetes type II and AIDS. In model organisms like yeast, mouse and E. coli, α-Gls has been used in many studies.17–21 As the concentration of human enzyme is scarce, the essential role of the enzyme of these model organisms has been bolded. Based on our results, the chimpanzee`s enzyme showed the closest relation to human counterpart and might always be the best substitute. However, the yeast and E.coli α-Gls seemed to be structurally different from the mammalians one and could not be considered as a suitable model. Based on the evolutionary point of view, it seemed that chimpanzees and humans share the most identical sequences and domains, and may have a common ancestor which a long time ago diverged from the mouse and rat ancestor. The plants α-Gls was significantly different from the mammalians one since they lack the glucoamylase site. However, the homology of their maltase site was quite noticeable. Therefore, as depicted in (Figure 2), the eukaryotic enzyme had been diverged significantly from the prokaryotic α-Gls since the separation from the common ancestor.
Entry |
Block 1 |
Block 2 |
Block 3 |
Block 4 |
Block 5 |
Mammalian α-Gls |
185-LLTAEYQTSNRFHFKLTDQT |
236- PFSIKVTRRSNNRVLFDSSIGP |
292- NVYGLGEHVHQQYRHDMNWKTWP |
854- LFCKTLCMDAVQHWG |
900-KRSFILTRSTFAGSGKFAAHWLGDNTATW |
Table 1 Conservative blocks of mammalian α-Gls
Entry |
Block 1 |
Block 2 |
Block 3 |
Block 4 |
Block 5 |
Mammalian Glucoamylase |
1314- EKIDCYPDENGAS |
1361- QYNSHGATADISLK |
3250- IIWDSQLLGFTFSDMFIRISTRLP |
3305- PPGYKKNSYGVHPYYMGLEEDGSAHG |
3355- GGILDFYVFLGPTPEIVTQQYTELIGRPVMVPYWSLGFQLCRYGY |
Table 2 Conservative blocks of mammalian glucoamylase
None.
The author declares no conflict of interest.

©2015 Mehraban, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.