MOJ
eISSN: 2573-2919


Research Article Volume 3 Issue 3
1Department of Agronomy, Assistant Professor, Lovely Professional University, India
2School of Biochemistry, Devi Ahilya Vishwavidyalaya, India
Correspondence: Prasann Kumar, Assistant Professor, Department of Agronomy, School of Agriculture, Lovely Professional University, Jalandhar, 144411, Punjab, India
Received: November 11, 2017 | Published: May 18, 2018
Citation: Kumar P, Pathak S. Nitric oxide: a key driver of signaling in plants. MOJ Eco Environ Sci. 2018;3(3):145-148. DOI: 10.15406/mojes.2018.03.00079
The colorless gaseous compound nitric oxide (NO) is a lipophilic free radical that diffuses readily through the plasma membrane. These attributes make nitric oxide ideal for a transient paracrine (between adjacent cells) and autocrine (within a single cell) signaling molecule. The half–life of NO in biological tissues is estimated to be < 6 Sec. This short half–life reflects the highly reactive nature of NO. It reacts directly with metal complexes and other radicals and indirectly as a reactive nitrogen oxide species with DNA, protein, and lipids. NO was first described in 1772 as nitrous air by Joseph Priestly the English theologian chemist and by co–educators. He was also the first to describe nitrous oxide (N2O), which he named nitrous air diminished. Priestly’ nitrous air induced a sensation of mild drunkenness, often coupled with bouts of uncontrollable laughter.
Keywords: agriculture, biotic, cadmium, diffuse, nitric oxide
Fast forward to the most recent time, NO has been demonstrated to orchestrate a plethora of physiological function in mammals, was the subject of the Nobel Prize in 1998 and was named molecule of the year in 1992 by journal of science. Despite of animal it was first reported in plant in 1779 by Klepper.1 Almost two decades later by the late 1990s, plant biologists had started to pay attention to NO playing role in plant immunity, initially in potato (Solanum tuberosum)2–4 and then two years later in Arabidopsis.5–7 However the Era is now gathering substantial momentum. It was undoubtedly an era of great enthusiasm for NO as a plant signaling molecule and today it is known to play a crucial role in the regulation of physiological process ranging from development to adaptation to biotic8–10 and abiotic stress.11 Furthering the application of new tools and technologies to study and addressing the molecular mechanisms employed by NO to control a variety of key cellular processes.12,13 Nitric oxide has a potent skill to encroach on a noticeable plant world due its enormous owndom like free radical, small size, no charge, short lived and permeability across biological membranes. It has also regulated signaling pathway for oxidative stress, mitochondrial activity, programmed cell death,14,15 growth and development in plants and responsible for the guidance of markable spectrum of plant cellular mechanism. The role of NO in plant may be equally diverse. The participation of NO in plant disease resistance pathways has been reported on several occasions5,6,16,17 and role of NO in the abscisic acid (ABA) signal transduction pathway leading to stomatal closure has been also demonstrated.18 The last decades, the role of No in tolerance of abiotic stress has established much consideration, it has begun to emerge as an important endogenous signaling molecule in the adaptation of plant to abiotic stress. As it is evident from the present review, recent progress on NO potentiality intolerance of plants to environmental stresses has been impressive.19 These investigations suggest that, NO it possesses antioxidant activity and might act as a signaling in activating ROI–scavenging enzyme activities under abiotic stress. NO responses to stress due to salt, drought, temperature, UV–B and heavy metal, which in their extreme limit responsible for cause of serious threats and set the plants with impaired growth, physiological and biological activities that are witnessed by lost in crop growth and yield etc. This review represents attention to the description of numerous NO sources, synthesis, the myriad role of NO and the molecular mechanism underpinning their function. Nonenzymatic NO production, according to the equation should occur only at pH below 4.5, since the pKa of nitrous acid is about 3.2.
NO is involved in plant metabolism and the nitrification and denitrification cycle provides NO as a byproduct of nitrous oxide oxidation into the environment by mean of a non–enzymatic mechanism.20 The studies of NO on plant metabolism date back to the 1960s when Fewson et al.21 address the recruitment of NO by microorganisms and higher plants. NO was suggested to be the key intermediate in the metabolism of inorganic nitrogen compound in higher plants and nitrogen fixing organisms. It was only in 1994 that NO was proved to be endogenously produced in a non–enzymatic way through conversion of nitrogen dioxide to NO by carotenoids in the light.22 Several plant systems use nitrite as a substrate, according to the basic reaction.
;
Moreover synthesis of NO on the apoplast has also been described by a non–enzymatic mechanism, whereby nitrite is converted to NO under acidic conditions in response toabscisic acid and gibberellins.23 These are also like Cytosolic NR (cNR), A plasma membrane–bound NR (PM–NR) associated with a PM– nitrite: NO reductase, Mitochondrial electron transport, Xanthine dehydrogenase/oxidase, Non–enzymatic NO formation at acidic pH. Nitric oxide is produced by a group of enzymes called nitric oxide syntheses. These enzymes convert arginine into citrulline, producing NO in the process. Oxygen and NADPH are necessary co–factors. 24 There are three isoforms of nitric oxide synthase (NOS) named according to their activity or the tissue type in which they were first described. The isoforms of NOS are neuronal NOS (or nNOS), endothelial NOS (or eNOS) and inducible NOS (or iNOS).25 These enzymes are also sometimes referred to by number, so that nNOS is known as NOS1, iNOS is known as NOS2 and eNOS is NOS.3.Despite the names of these enzymes, all three isoforms can be found in a variety of tissues and cell types (Figure 1).
Both enzymatic and non–enzymatic pathways have been described, but there is no consensus in sight on central source of NO in plant, and even less its regulation. NOS isoform may also present in the mitochondria, including a constitutive (cmtNOS) and inducible (imtNOS) mitrochandrial NOS26,27 which are thought to derive from cytosolic nNOS and INOS, respectively. Some studies have failed to locate mtNOS isoform, which could be related to different experimental designs or methods utilized in the NOS activity assays.NOS proteins catalyze the NADP–dependent oxidation of arginine (Arg) to NO and Citrulline. However, genes encoding a structurally related enzyme have not been identified in higher plants despite the completion of numerous genome projects. Oxidative mechanisms include the production of NO from L–arginine (L–Arg), polyamines or hydroxylamines. By contrast, reductive routes are dependent upon nitrite as the primary substrate and include reduction via NADPH nitrate reductase (NR), a cytosolic enzyme associated with nitrogen assimilation, whose primary function is the reduction of nitrate to nitrite28 it can further reduce nitrite to NO by the mitochondrial electron transport–dependent reductase,29 Which uses arginine as a substrate.
There are three different isoforms of the NOS which classify on their localization Zhou et al.30 All NOS are active as homodimers converting L–arginine to L–citrulline and NO.
When the availability of L–arginine is reduced, these enzymes also produce superoxide anion and NO, which may create peroxynitrite. Also a loss of function mutant, no overproducer 1(nox1) has been reported to have several fold greater concentration of L–arginine and this plant line exhibits excessive NO and citrulline accumulation. These data therefore imply the existence of a plant NOS like enzyme. Further, numerous studies in both Arabidopsis and tobacco (Nicotiana tobacum) have implicated, such NOS like activities as the source of reactive nitrogen intermediates during the nitrosative burst associated with the plant immune function.5,6 A recent paper reported the presence of NOS in a single celled green algae Osteroccocus tauri. This algae NOS possessed 45% similarity to human NOS. This enzyme exhibited NOS activity in vitro and possessed similar property to animal NOS proteins in the terms of the km for L–arginine (12microM) and the rate of NADPH oxidation. Unfortunately, these genes to date, no direct ortholog seem to be present in Arabidopsis and higher plants.31 In the late 1990s, the palate of available tools to dissect NOS production was composed by several macrophagic NOS compound inhibitors such as NGmonomethyl L–arginine (NMMA) and arginine analogs, and assay for arginine to citrulline conversion were used to detect the presence of NOS like enzymes in the different plant tissues (e.g. root, leaves, stems) and organelles (e.g. peroxisome).6,17,32 Interestingly, increase the concentration of polyamines, spermine and spermidine induce NO release, but the actual reaction mechanism still not resolved. Polyamines mediated NO production has been thought to be involved in root development and embryogenesis33 cadmium toxicity34 and drought stress.35 Hydroxylamine mediated NO synthesis is also a potent source of NO synthesis while location still unclear. Hydroxylamine and Reactive oxygen intermediate is the substrate for NO synthesis. According to given hypothesis this pathway involved in regulation of ROI concentration, especially during reoxygenation of anoxic tissues.
Nitrate reductase located in the cytosol, which catalyzes reduction of nitrate to nitrite is encoded by two genes in Arabidopsis designated nitrate reductase NADH 1 (NIA1) and NIA2 with NIA2encoding the enzyme which is responsible for NR activity. This enzyme has also potential to catalyze the reduction of nitrite to NO.28,29 Various reports provide knowledge in the respect of role for NR in the generation of NO in various cellular processes respectively, stomatal closure, osmotic stress; the plant defence response and auxin reduced lateral root formation.36 A plasma membrane–bound NiNOR activity was first described in tobacco, with activity being limited to the roots. The nitrite as substrate for NiNOR is probably provided by plasma membrane–bound NR in a coupled reaction. This enzyme generates extracellular NO and has been suggested to play a role in the sensing nitrate availability and during interactions with mycorrhizal fungi. Unfortunately, the identity of NiNOR still remains to be determined. NO can also be generated by nitrite reduction in the mitochondrial inner membrane, probably via cytochrome c oxidase and/or reductase.37 However, this only occurs when the oxygen concentration drops below 20 mM.29 NAD(P)H provides electrons via ubiquinone and the mitochondrial electron transport chain. This process has also been reported to produce small amounts of ATP. The peroxisomal enzyme xanthine oxidoreductase (XOR) can also reduce nitrite to NO. XOR has been shown to reduce nitrite to NO, using NADH or xanthine as the reducing substrate. However, this reaction only occurs under anaerobic conditions. As peroxisomes are a major site for the generation of ROIs, this organelle may provide an important location for the interaction of these species with RNIs32 (Figure 2).
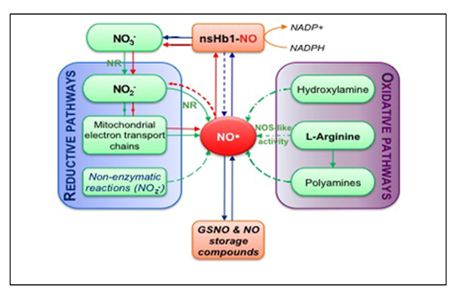
Figure 2 Simplified overview of NO biosynthesis and homeostasis in plant.
Source: Arc et al.42
Numerous genomic and proteomic methods have been spontaneously used to explicate meticulously NO dependent processes. Principle signal mechanisms convey their bioactivity between covalent modification of NO and RNIs with specific atoms of target proteins.38 The post translation modification process of S–nitrolylation, a redox modification of cysteine thiol group by NO exhibit a low pka sulfhydral group which supports significant susceptibility to a range of redox based post translational modification. The modification of these highly reactive Cys residue by NO and related RNIs are reversible except for sulfonic acid formation, the most highly oxidized modification (Figure 3). An experiment has set up to identify protein is regulated by S–nitrosylation in potato tissues. In this experiment, modified and optimize biotin switch assay and nano liquid chromatography combined with mass spectrometry was applied. This modified method provides a new dimension to better understand the signal transduction pathway to derive by NO transient signal. The first in planta biological function for S–nitrosylaltion emerged through a genetics approach, which uncovered a central role for SNOs in plant disease resistance. The exogenous addition of NO donors to plant protein extracts also demonstrated the in vitro formation of plant SNOs.39 The list of S–nitrosylated plant proteins are currently growing exponentially through the judicious application of the biotin–switch technique. For example, proteins specifically S–nitrosylated during plant immune function,9 cold treatment, heavy metal exposure and salt stress have been described. Unfortunately, current strategies for the identification of Cys redox switches on a global scale are not straightforward and typically lack sensitivity. However, new techniques are evolving to help achieve this.40 Another pressing current limitation in this area is the sensitivity of the biotin–switch and associated mass spectrometry methodology. In an interesting way, a unique prototype of NOS inhibitor was designed, known by Nanoshutter (NSI), which target the NADPH site of NOS and produces a specific florescence enhancement upon binding to constitutive NOS. The authors proposed that NSI is a promising tool with two photon excitation in the 800–9500 nm range 9.41 S–nitrosylated proteins are being identified at an increasing rate; deep insights into how these modifications might regulate protein function at the angstrom level are only just beginning to be obtained within a plant biology context. A primer for these studies was the recent demonstration of how S–nitrosylation of an NADPH oxidase, Respiratory burst oxidase homolog D (RBOHD), modulates the function of this key enzyme.10 Therefore, increasingly, NO–oriented research programs may need to embrace structural biology–based approaches.
Nitrate reductase located in the cytosol, which catalyzes reduction of nitrate to nitrite is encoded by two genes in Arabidopsis designated nitrate reductase NADH 1 (NIA1) and NIA2 with NIA2encoding the enzyme which is responsible for NR activity.
Authors are thankful to Department of Agronomy, School of Agricultures, Lovely Professional University, Jalandhar, for providing the consistent encouragement and undivided attention to the authors.
Author declares there is no conflict of interest.

©2018 Kumar, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.