Journal of
eISSN: 2574-8114


Review Article Volume 5 Issue 3
1University of Manouba, ISBST, BVBGR-LR11ES31, Biotechpole of SidiThabet, Ariana, Tunisia
2Laboratory of Microorganisms and Active Biomolecules, MBA-LR03ES03, Faculty of Sciences of Tunis, University of Tunis El Manar, Tunis, Tunisia
Correspondence: Mohamed Neifar, University of Manouba, ISBST, BVBGR-LR11ES31, Biotechpole of SidiThabet, Ariana, Tunisia
Received: April 23, 2019 | Published: May 10, 2019
Citation: Sghaier I, Guembri M, Chouchane H, et al. Recent advances in textile wastewater treatment using microbial consortia. J Textile Eng Fashion Technol. 2019;5(3):134-146. DOI: 10.15406/jteft.2019.05.00194
The textile wastewaters (TWWs) are one of the major sources of environmental pollution, due to the presence of various recalcitrant dyes. It is estimated that about 300,000 t of synthetic dyes are discharged in TWWs every year worldwide. Thus, untreated or incompletely treated TWWs cause harm to aquatic and terrestrial life. To avoid the negative impacts associated to the discharge of TWWs into the natural ecosystems, effective dye remediation processes are being developed. Current methods of removing dyes from TWWs are generally regarded to be complex, expensive and energy demanding processes. Therefore, bioremediation of TWWs using microbial consortia has appeared as an emerging alternative for textile dyes removal. This chapter provides an updated literature on the application of microbial consortia in the treatment of TWWs, focusing on the mechanisms involved in dye biodegradation and the main interactions established between the consortia members and how they can influence dye removal efficiencies.
Keywords: textile wastewaters, bioremediation, co-cultivated microorganisms, azo-dyes
During the past few decades, water resources are getting scarcer due to exponential increase in population, agriculture, urbanization and industrialization.1–4 Different industrial sectors entail significant environmental and public health concerns. One such industry is textile dyeing which is one of the most water-intensive industries and generates releases consisting of recalcitrant organic molecules generally, problems of color, high concentrations of Biological Oxygen Demand (BOD), Chemical Oxygen Demand (COD), fibers, surfactants, detergents and solvents.5 Textile industries consume huge volumes of freshwater for its various wet processes and release equal amounts of wastewaters.6 During the dyeing process, not all the dyes are fixed to the fabrics. There is always a portion of unfixed dye which is discharged into the wastewater that forms the major pollutant in this effluent. Textile market utilize more than half of world dye and organic pigment, and the demand is expected to increase more than $30 billion in 2019.7 Different types of dyes are used in textile industries, the most commonly frequent dyes applied in dyeing units are azo dyes. Apart from textile industry, they are also used in tannery, paper and pulp, pharmaceutical, food, paint, plastics, cosmetics and electroplating industries.8 The improper discharge of colored dye effluents into natural water bodies severely affects all living forms and causes aesthetical unpleasantness creating a significant problem to human being.9,10 Removal of dyes from effluent has been given a top priority. Several physico-chemical methods have been employed but they have facing several problems, such as generation of toxic by-products and economical unfeasibility.10,11 Bioremediation has become a very special challenge since it is cost-effective, eco-friendly and does not produce a large quantity of sludge.12 Several studies have focused on the utilization of pure culture to decolorize synthetic dyes. Due to the chemical complexity of these dyes, it is necessary to develop more efficient microbial processes for decolorization.10 Recently, trend is shifting towards use of microbial consortia. Several microbial consortia have been reported for efficient dye removal.13 In this study the main aim is to emphasize on the existing literature on microbial decolorization of TWWs using co-cultivated microorganisms.
Characteristics of textile wastewaters
Textile dyeing industries are facing problems to meet the green practices standards for safe discharge of wastewater due to its complex nature. TWWs are a complex mixture of salts, acids, heavy metals, organ-chlorine-based pesticides, pigments and dyes.5,10 TWWs generated from the different wet processes are characterized by high pH, temperature, BOD, COD, detergents, surfactants, suspended and dissolved solids, dispersants, leveling agents, toxic organics, chlorinated compounds, sulphide and formaldehyde, may be added to improve dye adsorption onto the fibers9 and more details are mentioned in the Figure 1. Such effluents are also characterized by the presence of heavy metals, such as Cr, Zn, Cu and Al due to metal-based complexes dyes.10,14 The most common textile operates are desizing, bleaching, mercerizing, dyeing and finishing.15 Characteristics and the amount of TWWs depend to the process, dyeing is the most one which requires large volumes of water not only in the step of adding color to the fibers, in dye bath, but also during the rinsing step. Mercerizing and finishing are also significant generators of TWWs. In addition, equipment, machines and chemicals,5 such as detergents and stabilizers, alter significantly the nature of TWWs. Another important factor which contributes to the ecotoxicity and the volume of TWWs is that dyeing and finishing processes, especially, require the imput of a wide range of dyestuffs. The Variety of dyes depends to the fiber used.16 For example, cellulose fiber requires the application of direct, reactive, vat, azo or sulfide dyes. Acid dyes are used essentially for wool and silk. Azo and disperse dyes are applied to the polyester fiber. A large quantity of these dyes is released in the TWWs due to their degree of fixation to fibers.
Textile dyes and classification
According to Witt theory, dyes have a chromophore group which imparts color to the dye and auxochromes to intensify the color when introduced into a colored molecule. The most important auxochromes are amine (–NH3), carboxyl (–COOH), sulfonate (–SO3H) and hydroxyl (-OH).17 Based on the method of application, main classes of dyes are: acid, basic, direct, reactive, disperse, vat, mordant and sulphur18,19 and according to the chemical structure and the type of the chromophore present in the molecule, the most important classes, as indicated in Table 1, are azo, anthraquinone, nitro and other dyes. Azo dyes account for over 50% of commercial dyes available in the market and constitute the most important class applied in textile processing industries.14 The first azo dye, Aniline Yellow was reported in 1858 by Griss. More than 10,000 dyes are available commercially and more than 7×105 tons of dyestuffs are produced annually.9,20 Azo dyes are characterized by the presence of one or more azo groups (R1-N=N-R2), the persistence and the stability against microbial attack.
Class |
Chromophore |
Examples |
Applications |
Azo |
Methyl Orange |
Textile |
|
Anthraquinone |
Remazol Brilliant |
Textile |
|
Phthalocyanine |
Direct blue 86 |
Textile |
|
Nitro |
Acid yellow 1 |
Wool |
|
Silk |
|||
Sulfur |
Sulfur yellow 4 |
Cotton |
|
|
Table 1 Classification of dyes based on the chromophore group
The release of TWWs causes dark coloration of surface waters that captures the attention of both the public and the authorities. During textile processing, a large amount of dyestuff is liberated directly in the effluent due to inefficiencies in dyeing step. In addition, dyes absorb light in the visible range, being detectable even at a concentration of 1 ppm.14 Apart from the aesthetic unpleasantness, the main environmental concern with the dyes is their absorption and reflection of sunlight penetration, which, in turn, decreases photosynthetic activity and dissolved oxygen amount in the water, and affects aquatic flora and fauna.10,21,22 Several reports have indicated that direct and indirect toxic effects of the dyes and TWWs can cause tumors, cancers and allergies in humans.10,23,24 Due to their persistence nature, azo dyes have negative impact on the environment in terms of total organic carbon (TOC), COD and BOD.25 Many synthetic dyes and their metabolic intermediate products are found to be toxic, mutagenic and carcinogenic.26,27 The Major toxic effects of azo dyes are caused by aromatic amines generated after their biodegradation. Also, there are a large group of aromatic amines which are either cancer-suspect agents or established mutagens in the standard Salmonella mutagenicity assay.28 Some azo dyes can be carcinogenic without being cleaved into aromatic amines. However, the carcinogenicity of many azo dyes is due to their cleaved product such as benzidine. Benzidine induces different human and animal tumors. Another azo dye component p-phenylenediamine (p-PDA) is a contact allergen.29 Reports are also available on other important dyes including triphenylmethane and anthraquinone which have been shown negative impacts.12,30,31 Some dyes from the triphenylmethane group have been reported to be toxic, mutagenic, and carcinogenic.30,31 More details about impacts of TWWs are indicated in Figure 1.
Considering the several hazards of dyes, their removal before their discharge into surrounding environment is urgent. So, many technologies have been developed to achieve an efficient and economic treatment. They can be physical, chemical or biological.
Physico-chemical methods
During the past two decades, several physicochemical decolorization techniques, such as coagulation-flocculation, adsorption, oxidation and membrane techniques, have been reported.32 Physical methods are mainly used for the primary treatment of TWWs.20 Based on coagulation–flocculation of dyes, they are effective for the removal of mainly sulphur and disperse dyes, but show very low capacity for acid, direct, reactive and vat dyes.21 Although activated carbon is a very effective adsorbent for various types of dyes, it is not often used due to its high cost and removal pH dependent but it is capable of treating only small effluent volume, operates at slow speed.33 For Chemical treatments, excess use of chemicals requires high cost and results into secondary pollution.34 These methods can only transfer the dyestuff from one phase to another leaving the problem essentially unsolved. Briefly, their continuous application put forth their limitations as it is detailed in Figure 2, giving the relay to biological treatment.
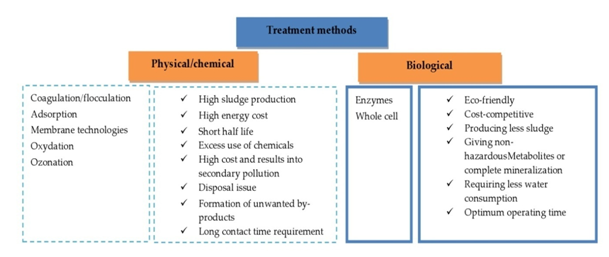
Figure 2 Different treatment methods of textile wastewaters and importance of biological treatment reference to conventional techniques.
Biological treatment
Biological processes provide an alternative to existing technologies for many purposes (Figure 2). Several microbial sources have been reported including fungi, bacteria, yeasts, algae and actinomycetes.21 Extremophilic microorganisms are widely applied in TWWs treatment due to their ability to survive in such harsh condition which poses a limiting factor for mesophilic microorganisms.35,36 Plant growth promoting rhizobacteria show also interesting ability to decolorize various textile dyes.37,38 These bioprocesses can be classified, on the basis of oxygen requirement, into aerobic, anaerobic and anoxic or facultative or a combination of these.39 And they are based on the enzymes synthesized by microorganisms40,41 or whole cells. The effectiveness of microbial decolorization depends on the adaptability and the activity of selected microorganisms enhanced by its application under optimal conditions.42 This decolorization can be achieved through individual and specific strain or co-cultivated microorganisms.
Pure culture
Bacterial pure culture: There are many published studies dealing with the use of pure cultures in decolorization process (Table 2). Therefore, a large number of microorganisms and enzymes have been isolated. Among these microorganisms, bacteria are frequently used. Attempts to isolate pure bacterial cultures capable of degrading azo dyes started way back in 1970s with isolation of Bacillus subtilis, Aeromonas hydrophila and Bacillus cereus.10,43 As reflected in Table 2, there are other extensive reports describing the decolorization of reactive azo dyes mediated by pure culture. Among these strains, Pseudomonasis largely exploited to decolorize commercial textile azo dyes, such as Remazol Orange 3R by Pseudomonas aeruginosa strain BCH with removal of 98 % within 15 minutes44 or Black B.45 Application of the genus Halomonas46,47 show also promising results azo dye degradation. A.hydrophila and some other aerobic bacteria were studied for azo dye decolorization under aerobic condition by oxygen insensitive or aerobic azoreductases, because azo reductase activity is inhibited in presence of oxygen.48
Fungal pure culture: Phanerochaete chrysosporium is the most frequent, robust and model white-rot fungus reported in the literature for decolorizing several dyes from TWWs.49,50 Also, three promising strains, Dichomitus squalens, Ischnoderma resinosum and Pleurotus calyptratus show an efficient decolorization of both Orange G and Remazol Brilliant Blue R.51 Another white rot fungus Irpex lacteus is capable to decolorize the chromium metal complex dye Isolan Dark Blue 2SGL-01.52 In addition, Trametes pubescens was found to be the most effective strain in terms of decolorization performance on the azo dye Congo Red, in submerged culture,53 Trametes maxima CPB3054 and Fomes fomentarius55 show an ability to remove Remazol Brilliant Blue R dye. However, degradation of dyes in TWWs by white-rot fungi has some intrinsic drawbacks like the long growth phase and the requirement of nitrogen restrictive environments, unreliable enzyme production and large reactor size due to the long holding time for complete degradation.7,14,56 Multiple fungal species, particulary the genus Aspergillus described by57 are able to decolorize a large number of dyes. This genus was also reported by.58 Some recent examples are detailed in Table 2.
Yeast pure culture: Yeasts present significant advantages such as low cost, readily availability of biomass source and resistance to extreme environment conditions. Furthermore, several yeasts have been found capable to treat TWWs.59The genus Candida is widely used in dye decolorization and through biodegradation mechanism60–62 or biosorption63,64 or also by bioaugmentation.65 Thermo-tolerant yeast, Kluyveromyces marxianus IMB3C has shown the ability to remove Remazol Black-B.66
Importance of treatment with microbial consortium reference to treatment with mono-culture: Due to the limitations of pure strains on decolorization, such as the narrow range for decolorization of different azo dyes and the inability to degrade completely or mineralize these dyes,67 it has been observed that microbial consortia are mainly beneficial as they can conjointly carry out degradation tasks that no single culture can begin effectively.14,68–70 In addition, reports considered the consortium as a suitable combination of efficient strains which can be used in bioremediation of TWWs providing a rich metabolic network.71 In a mixed culture system, the degree of biodegradation and mineralization of dyes is higher due to the synergism of metabolic activities of a microbial community.7,72,73 In microbial consortium, the individual strains may attack the dye molecule at different positions or may utilize metabolites produced by the co-existing strains for further decomposition.68 In such approaches microbes acclimatize themselves to the toxic wastes and new resistant strains develop naturally, which then transform various toxic chemicals into less harmful forms.19,74 The complete degradation of chemical substances is only possible in the presence of several enzymes produced by the mixed cultures.14,75 Aromatic amines generated after cleavage of azo bonds are often toxic in nature. But, in a mixed culture, these aromatic amines can be degraded by the synergistic action of organisms.76,77 76reported three isolates Micrococcus sp., M. luteus, and P. polymyxa, when used in mixed culture were able to decolorize nine dyes but individually used, they were found to be inefficient for dye removal.14,76
Bacterial consortium: Bacterial consortia are the most frequently used for decolorization of azo dyes, as they are generally fast to multiply rapidly under aerobic, anaerobic, facultative conditions as well as in extreme environmental conditions, like high salinity and wide variations in both pH and temperature.14,22 The efficiency of bacterial consortium decolorization compared to pure culture removal is may be due to the involvement of quorum sensing, mechanism by which bacteria regulate gene expression in accordance with population density through the use of signal molecules. Quorum sensing allows bacteria populations to communicate and coordinate group behavior. Recent studies regarding the biodegradation of dyes in TWWs using bacterial consortia are reported in Table 2. The bacterial decolorization can be directly influenced by various factors78,79 such as the level of agitation, oxygen, temperature, pH, dye structure, dye concentration, supplementation of different carbon and nitrogen sources, electron donor, redox mediator80,81 and salt concentrations. Therefore, acclimatized bacteria, isolated from dye contaminated sites, are very efficient in removal process due to adaption to different extreme environmental conditions.14 Optimization of such abiotic conditions makes the microbial system more efficient and practicable (Table 3).
Strain |
Dye |
Concentration (mg L-1) |
% decolorization |
Time |
References |
Yeast culture |
|||||
Trichosporon beigelii |
Navy blue HER |
50 |
95% |
24 h |
(Dafale et al.25) |
Candida krusei |
Basic Violet 3 |
10 |
100% |
24 h |
(Deivasigamani and Das,82) |
Trichosporon akiyoshidainum HP2023 |
Reactive Black 5 |
300 |
100% |
24 h |
(Martorell et al.83) |
Candida tropicalis TL-F1 |
Acid Brilliant Scarlet GR |
100 |
100% |
24 h |
(Tannadal et al.61) |
Fungal culture |
|||||
Trichoderma tomentosum |
Acid Red 3 R |
85.5 |
99.20% |
72 h |
(He et al.84) |
white-rot fungus Cyathus bulleri |
Kiton blueA |
50 |
88% |
6 h |
(Vats and Mishra,85) |
Trametes versicolor CBR43 |
acid, disperse and reactive dyes |
200 |
90% |
9 days |
(Yang et al.86) |
Bjerkandera adusta OBR105 |
Reactive and acid dyes |
200 |
99% |
3 days |
(Sodaneath et al.87) |
Pleurotus sp. MAK-II |
Congo Red |
150 |
96% |
NA |
(Manavalan et al.88) |
Remazol Brilliant Blue R |
150 |
72% |
|||
Marasmius cladophyllus UMAS MS8 |
Remazol Brilliant Blue R |
200 |
100% |
15 days |
(Singh et al.89) |
Bacterial culture |
|||||
Pseudomonas extremorientalis BU118 |
Congo red |
100 |
75% |
24 h |
(Neifar et al.36) |
Nesterenkonia lacusekhoensis EMLA3 |
Methyl red |
50 |
97% |
16 h |
(Battacharaya et al.90) |
Serratia liquefaciens |
Azure-B |
100 |
90% |
48 h |
(Haq et al.91) |
Bacillus aryabhattai DC100 |
Coomassie Brilliant Blue G-250 |
150 |
100% |
72 h |
(Paz et al.92) |
Bacillus cereus RJVL 2514 |
Reactive Violet 13 and |
100 |
88% |
NA |
(Gangavarapu et Ravuri,93) |
Reactive Blue 171 |
87% |
||||
Halomonas sp. strain A55 |
Reactive Red 184 |
150 |
96% |
24 h |
(Guadie et al.47) |
Staphylococcus sp. |
Remazol |
100 |
100% |
12 h |
(Kharthikeyan et al.94) |
K2204 |
Brilliant Blue R |
||||
Table 2 Recent reports on microbial pure culture capable of dye degradation
NA, Not Available
Strain |
Dye and concentration (mg/L) |
Class |
Condition (Temp.(˚C), pH, agitaion) |
Decolorization (%) and time (h) |
References |
Bacillus flexusNBN2, Bacillus cereus AGP-03, Bacillus cytotoxicusNVH 391-98 and Bacillus sp. L10 |
Direct Blue 151 (DB151) |
Double Azo |
36, 9.5, NA |
87, 5days |
95 |
Direct Red 31 (DR 31) 200 |
|||||
Acinetobactersp. and Klebsiellasp. |
Reactive Orange 16 Reactive Green 19 |
Anionic single azo |
30, 7, static |
80, 72 |
96 |
Double azo |
|||||
Bacillus sp., Staphylococcus sp., |
Remazol Brilliant Violet 5R (RBV5R),200 |
Azo |
37, 6.5, microaerophilic |
100,18 |
73 |
Escherichia sp., Enterococcus sp. and Pseudomonas sp. |
|||||
M1C (highest similarity to Zobellellataiwanensisstrain AT 1 and M2C (Bacillus pumilusstrain HKG212) |
Reactive green-19, 100 |
Azo |
32, 8.3, NA |
97, 24 |
97 |
Microbacteriumsp., Leucobacteralbus, Klebsiellasp. and Staphylococcus arlettae |
Disperse Red 1 |
Azo |
36, 7, anaerobic–aerobic reactor |
80, 72 |
98 |
15 different bacteria |
Trypan Blue, 50 |
Double azo |
30, 7, NA |
100, 24 |
99 |
Consortium GR: Proteus vulgaris and Micrococcus glutamicus |
Green HE4BD and many other reactive dyes(Golden Yellow HE4R, Orange 3R, Violet 5R, Red ME4BL and Red M2BN), 50 each) |
Reactive double azo (Reactive double azo, Reactive single azo) |
37, 8, static, anoxic |
100, 24 |
68 |
Bacillus vallismortis, Bacillus pumilus, Bacillus cereus, Bacillus subtilisand Bacillus megaterium |
Congo red, 10 |
Double azo |
37, NA, 130 rpm |
71, 72 |
100 |
Salmonella subterranea and Paenibacilluspolymyxa |
Reactive Blue 4, 300 |
Anthraquinone |
35, NA, NA |
24, 24 h |
101 |
Pseudomonas pseudoalcaligenes, Pseudomonas citronellolis and Pseudomonas testosterone |
Acid Orange 7, 100 |
Azo |
37, NA, aerobic |
90, 24 |
102 |
Pseudomonas aeruginosa, Bacillus flexusand Staphylococcus lentus |
Acid blue 113, 800 |
Double azo |
37, 100 rpm, isothermicbioreaction calorimeter |
93.7, 12-16 |
103 |
Pseudomonas putida and Shewanellaoneidensis |
Congo red, 0.5 mM |
Double azo |
NA |
100, 72 |
104 |
Twelve acclimatised consortia |
Reactive Violet 5R, 100 |
Single azo |
90, 30, shaking and static |
100, 30 |
74 |
Consortium : (aerobic granular sludge) |
Reactive |
Single azo |
30, 140 rpm, microaerophilic |
100, 80 |
105 |
Yellow, 15.5 |
|||||
Consortium of Purple Non-Sulfur Bacteria (PNSB) |
Reactive Red 159,500 |
Azo |
Anaerobic sequencing batch reactor (ANSBR) |
97.68 ± 0.74 |
106 |
Pseudomonas, Arthrobacter, and Rhizobium |
Acid Orange 7, 200 |
Azo |
28±2, 6.25±0.25, agitation, Packed-Bed BAC reactor |
90, 96 |
107 |
Actinobacteria, Bacteroidetes, Proteobacteriaand Firmicutes |
Acid Brilliant Scarlet GR, 100 |
Azo |
30, 7, 150 r/min, Aerobic, batch processes |
100, 12 |
61 |
Consortium (TJ1), Aeromonascaviae, Proteus mirabilis and Rhodococcusgloberulus |
Acid Orange 7, 200 |
Azo |
37, 7, microaerophilic |
90,16 |
108 |
Providencia sp. SDSandPseudomonas aeuroginosa strain BCH |
A mixture of reactive azo dyes: Red HE3B, Red HE8B, and Remazol Blue. |
Azo |
30, 7 |
100, 20 |
109 |
consortium BDN: Alcaligenes sp., Bacillus sp., Escherichia sp., Pseudomonas sp., Providencia sp., Acinetobacter sp., Bacillus sp. and Bacillus sp. |
Mixture of reactive azo dyes, 300 |
Azo |
Microaerophilic fixed film reactor |
100, 24 |
72 |
Halophilic consortium VN.1: Pseudomonas fluorescens, Enterobacteraerogenes, Shewanella sp., Arthrobacternicotianae, Bacillus beijingensisand Pseudomonas aeruginosa |
Reactive Blue 220 (RB220), 2,500 |
Formazan |
30, 8, static, 7% NaCl |
100, 30 |
110 |
Pseudomonas sp., Brevibacillus sp. and two strains of Stenotrophomonas sp |
dye mixture, 100 |
|
37, 6.5, static, microaerophilic |
94, 24 |
71 |
Table 3 Decolorization of various azo dyes by bacterial consortia
Other microbial consortia
Conversely to decolorization by bacterial consortium, few reports are available on decolorization of dyes by yeast consortia and fungal consortia, especially on decolorization by biodegradation mechanism not by biosorption or bioaugmentation. Another approach is the synergistic action of fungal-bacterial consortium which provides an alternate way for efficient removal of various contaminants. Some reports indicated that fungi-bacteria co-cultures showed higher efficiency and stability111–114 to degrade the mixture of many kinds of macromolecule organics into small-molecule substances which can be further degraded or even mineralized by bacteria.65,115 In addition, few studies illustrate the ability of bacterium yeast consortium to treat TWWs.116 Apart from these approaches, A synergistic strategy occurred by involving plants such a plant-bacterial system using Glandularia pulchella and Pseudomonas monteilii ANK117 or plant consortia.118,119
It is very much important to understand well the mechanism by which azo dye decolorization is realized. Generally, biodegradation of azo dyes is achieved via two mechanisms either direct decolorization or indirect/mediated decolorization. Behind both mechanisms, enzymes are well involved to degrade recalcitrant compounds in the microbial system.120 Several reports demonstrate the degradation of complex organic substances by different enzymes including laccase,121,122 azoreductase,123,124 lignin peroxidase,6 NADH-DCIP reductase99 and hexane oxidase.25 Among these families of enzymes, laccases and azoreductases have shown a great potential to decolorize a large range of known industrial dyes. Biodegradation of mainly azo dyes is due to the reduction of azo bonds (-N=N-) by azo-reductase enzymes under anaerobic condition.14 During this process, four electrons are transferred from electron donors to the electron acceptor (azo dye) in two stages at the azo linkage, resulting in dye decolorization and generation of colorless amines.10 Generated metabolites or aromatic amines are then further degraded aerobically or anaerobically. The role of laccase in asymmetric cleavage of azo dye was well reported in biodegradation of anthroquinone and azo dyes by a fungal strain Lentinus sp.125 The laccases from Brevibacillus sp. (Z1)126 and Anoxybacillus Gonensis P39127 which are applied in removal of various azo dyes were purified and characterized. Under aerobic conditions, respiration may dominate utilization of NADH which inhibits the electron transfer from NADH to azo linkage. Thus, decolorization might be related to non-specific extracellular reactions occurring between reduced compounds generated by the anaerobic biomass.10 In anaerobic conditions, a low redox potential (≤50mV) causes the effective decolorization of the azo dyes decolorization and azoreductases were found to be oxygen sensitive and released extracellularly.128 In order to stimulate the removal process of dyes, researchers use redox mediators. Numerous studies proved that addition of redox mediators, such as synthetic electron carriers, anthraquinone-2-sulfonate or quinone compounds enhance significantly biodegradation of azo dyes. However, the high cost of quinone addition limited its usage. To make practical the application of bacteria-mediated decolorization, other redox mediators like activated carbon, carbon black and carbon nano tube were used.
Many analytical techniques are applied to identify the intermediates and aromatic amines generated during azo dye decolorization. UV-Vis analysis is a preliminary method by which products produced during biodegradation after incubation under anaerobic conditions were studied by following the change in the ultraviolet visible (UV-Vis) spectra. The dye decolorization was estimated by measuring the absorbance at λ max of the dye using an UV-Vis spectrophotometer.129 For further detection of metabolites, Thin Layer Chromatography (TLC) and High Performance Thin Layer Chromatography (HPTLC) techniques are used.130 These methods give insights in numbers and types of metabolites generated. But for confirmation of the degradation of dye, HPLC has been largely used.10 Moreover, Fourier Transform Infrared Spectroscopy (FTIR) is also widely applied to check the removal of azo group from the azo dye and generation of new type of compounds.131 Another powerful techniques used for qualitative determination of metabolites are Gas Chromatography-Mass Spectrometry (GC-MS) and Liquid Chromatography-Mass Spectrometry (LC-MS). The evaluation is occurred via molecular weights and structural information which is used further to propose the degradation pathway of the dye. At present, apart from these methods, advance technique, Nuclear Magnetic Resonance (NMR) is applied to get detailed quantitative study.132
Microbial degradation of azo dyes generates intermediate organic compounds, aromatic amines, which are frequently reported to be mutagens and carcinogens. By-products generated may sometimes be even more toxic than the dyes themselves.21 Therefore, it is vital to evaluate the toxicity of both azo dyes and their metabolites. The toxicity caused by dye metabolites may not have the same effect on all levels of biological organizations. Basis on this fact, the studies on microbe-mediated dye detoxification are categorized into molecular, cellular, organismic and ecological levels.133 Molecular toxicity includes mutagenicity/genotoxicity which can be caused via oxidative DNA damage, micronuclei formation or chromosomal aberrations23 To determine this toxicological category, the widely used assays are Ames test, Comet assay,109 TUNEL assay and Allium cepa. Cellular level is the most investigated in the studies about toxic impacts on humans and organismic toxicity is used to identify toxicological impacts on model organisms including animals and plants. Conversely to these levels, ecotoxical studies are too complex. Thus, the main objective of ecotoxicity is to extrapolate and predict risk under laboratory conditions. More details about the different tests used for evaluation of toxicity azo dyes are mentioned in the Table 4. All reports98,103,109 using microbial consortium for TWWs treatment and degradation of azo dyes prove that it is an efficient approach to reduce significantly the toxicity of TWWs and azo dyes.140–150
Methods |
Organisms |
References |
Genotoxicity |
Allium cepa |
134 |
Mutagenicity |
Salmonella |
98 |
E. coli |
135 |
|
Cytotoxicity |
Allium cepa |
109 |
MTT assay with human cell lines of keratinocytes (HaCat) |
103 |
|
Human blood cells |
23 |
|
Oxidative stress |
Allium cepa |
109 |
Phytotoxicity |
Vigna radiata |
91 |
Triticum aestivum |
136 |
|
Phaseolus mungo |
136 |
|
Zeamays L. |
137 |
|
Sorghum vulgare |
138 |
|
Phaseolus aureus |
110 |
|
Lactuca sativa |
139 |
|
Acute toxicity |
Daphnia similis |
98 |
Hydra attenuata |
98 |
|
Daphnia magna |
99 |
Table 4 Methods used for evaluation of toxicity of azo dyes
TWWs pose a significant threat to the environment due to long term disposal. Physico-chemical techniques failed to decolorize TWWs in an efficient and economical manner. Bioremediation has been emerging approaches to tackle azo dye pollution. The overall evaluation of biological treatment suggests a positive outlook for microorganisms as a potential candidate for the removal of dyes from TWWs.
In this approach, co-cultivated microorganisms prove a significant efficiency in dye decolorization. Microbial consortia was found to be advantageous than pure cultures due to concerted metabolic activities of microbial community.
Although there are a large number of studies focusing on the field of dye decolorization, few studies75 have targeted the real effluent or TWWs treatment reference to azo dyes removal. Thus, more attention should be paid to commercialize and apply microbial consortia in TWWs treatment.
The authors acknowledge the financial support from the Tunisian Ministry of Higher Education and Scientific Research in the ambit of the laboratories’ Projects BVBGR-LR11ES31 and MBA-LR03ES03.

©2019 Sghaier, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.