Journal of
eISSN: 2574-8114


Research Article Volume 5 Issue 2
1Microbiology Division, Biological Material Laboratory, CSIR-CLRI, India
2Department of Textile Technology, Anna University, India
Correspondence: Dr. A. Gnanamani, Microbiology Division, Central Leather Research Institute (CSIR-New Delhi), Adyar, Chennai 20, Tamil Nadu, India, Fax 91-44-24912150
Received: June 19, 2017 | Published: March 6, 2019
Citation: Iswarya S, Shanuja SK, Giri Dev VR, et al. Asperyellone – a suitable coloring agent for protein based textile fabrics: an approach on production, characterization and application. J Textile Eng Fashion Technol. 2019;5(2):73-79. DOI: 10.15406/jteft.2019.05.00184
Color is the first thing which is perceived by the senses. Natural colorants are extremely safe because of its added advantages. The present study describes extraction, characterization and application of a yellow pigment from Aspergillus species, strains AN01 and explores its wider applications. The chosen strain isolated from the soil displayed an extracellular yellow pigment production when grown at specific nutrient and growth conditions. The yellow color pigmented solution free from spores extracted, concentrated, purified and subjected to characterization using UV, FT-IR, DSC, TGA, GC-MS and NMR analyses. From the results the yellow pigment is identified as Asperyellone. Coloring efficacy of the yellow pigment on silk, cotton, synthetic and wool fabrics demonstrates appreciable color uptake by the materials. Increased dye uptake is observed in the protein fibers and polypropylene. The high K/S values corroborate substantiates the preference of Asperyellone pigment to proteins than other natural or synthetic fabrics. The fastness of the pigment on the various substrates is found to be good except to the light fastness. Additional molecular docking studies with silk protein and Asperyellone display the interaction and appreciable binding energy. The above results authenticate the suitability of yellow pigment to enhance the organoleptic property of silk, which ultimately will improve the export value at appreciable level.
Keywords: colorants, Aspergillus, asperyellone, dye uptake, fibres, molecular docking
Color imparts attraction to the marketable products like food, fabrics and pharmaceutical products, etc. Colorants obtained from natural sources like plants, animals and microbes are termed as Biocolorants. Bio-colorants offers various advantages over synthetic colorants like non-toxic, biodegradable and ecofriendly nature.1 Synthetic colorants are manufactured from various hazardous chemicals which leads to harmful effects on workers, consumers and finally to the environment.2 Textile industries are one among the major industries which rely on the use of synthetic colorants. Synthetic dyes have the ability to color the fabric using simple methods and in addition it renders good fastness properties compared to that of natural colorants. The faster acceptability of the synthetic colorants lies in its wide color range and long lasting nature. Wastewater from textile industry contains heavy loads of toxic pollutants like dyes, heavy metals which are harmful to the environment.3 Microbial dyes or pigments are very good alternative to this issue because of its added advantages like fast growth in cheap medium, availability, independent from weather conditions and reproducibility. In addition to its clinical effects like antioxidant, antiinflammatory and antimicrobial activity, the shades produced by microbes are biodegradable and ecofriendly4 of that, Microbes, particularly fungi produces large variety of stable pigments like anthraquinones, carotenoids, rubramines etc. Several studies have been reported on fungal pigments as colorants in textiles as well as food industry.5,6 Toxicity studies on fungal pigments on human skin revealed the biocompatibility nature of the pigments.
With regard to Apergillus family, they have been well recognized for the production of a wide variety of stable pigments like Fumonisms, Napthogamma pyrones, Biocoumarins, Malformins and Asperazines. More than 145 pigments from this family have been characterized with structural elucidation.7 Aspergillus niger produced several yellow pigments which have the structure of dimeric linear naphtha γ-pyrones.8 Ehrlich et al.9 isolated Fonsecin monomethyl ether as a natural metabolite of Aspergillus niger and suggested that the compound is the precursor of Aurasperones B and C. A brown pigment produced by the species of Aspergillus family demonstrated application in textile industry.10 Yu et al.11 reported the production of Asperyellone from both Aspergillus niger and Aspergillus awamorii during 1967. However, in the same period, a report on Asperenone from A.niger made by Jefferson.12 And during the year 1969 report on Asperenone from A. niger and A. awamorii was made by Gerald Pattenden.13 The yellow pigment production from Phellinus pini reported for its antifungal activity reported during 1996.14 Rao et al.15 reported the enzyme (15- Lipoxygenase) inhibitory and human platelet aggregation activity of asperenone isolated from Aspergillus niger during 2002.15 All the summarized information suggested that no reports are available on the application of Asperyellone after the year 2002. In the preliminary study, the compound showed very good coloring property, research initiatives are made to study the textile dyeing property of Asperyellone from A. niger and this is the first report on the textile dyeing property of Asperyellone.
Thus, the present study explores the extraction of a yellow pigment from Aspergillus species and studied the coloring property. In brief, the study details the production, extraction, characterization, identification, structural elucidation and application of a yellow pigment as a natural colorant in various fabrics.
Isolation of fungus
The fungus was isolated from the soil sample collected from CLRI premises, serially diluted and plated on Czapexdox (CPD) agar plates and incubated at 28˚C for 3-4 days for fungal growth. Colonies were confirmed as Aspergillus by lactophenol cotton blue staining technique and named as Aspergillus species strain AN01.
18S rDNA identification of the Aspergillus species strain AN01
The fungal DNA was isolated by method proposed with some modifications.16 PCR Amplification of the 18S rDNA carried out using the following primers. (Forward primer: GGGCTCCTTGGTGAATCATA. Tm 59.89, Reverse primer: CTCCGGAATCGAACCCTAAT. Tm 60.28) according to the method reported.17 Analysis of the PCR products was performed by electrophoresis on 1.5% agarose gel and the product was extracted from the gel and submitted for sequencing. The sequenced product was compared with some Aspergillus sequence from NCBI database and based on the similarity the species was identified. The identified Aspergillus species strain was submitted to NCBI for the accession number Genbank KY205487.
Growth, extraction and purification of a yellow pigment
Aspergillus species strain AN01 was cultured in CPD broth in 500ml Erylenmeyer flask for 5 days at 28˚C under shaking condition. The yellow pigmented biomass was separated and then extracted with acetone till the biomass become colorless. The acetone free yellow pigment was concentrated after subjected to rota-vapor evaporation and then stored at 4˚C. Purification of the pigment was carried out using activated silica gel (100-200 mesh) in a glass column. The mobile phase contain benzene: acetic acid: water at 4:1:1 ratio.8
Characterization of the pigment
UV-visible spectral analysis
UV-Visible spectral analysis of the sample was carried out using Shimadzu spectrophotometer UV 2450 (Japan), in the range of 200-800nm at 1nm resolution. Spectrum was taken after dissolving the samples in DMSO.
Fourier transform infra-red (FT-IR) spectrum
Fourier Transform infrared (FT-IR) spectrum was recorded in spectrum-I FT-IR spectrophotometer (Perkin-Elmer Co, USA.). In brief, 20µg of sample was mixed thoroughly with 200µg of potassium bromide (Sigma, USA) and pelletized using hydraulic press. FT-IR spectrum was measured range of 4000-400cm-1 at a resolution of 4cm-1. All measurements were of 20 scans with potassium bromide as background reference.
Differential scanning calorimetric (DSC)
Approximately, 10-20µg of sample was loaded in an aluminium DSC pan, gravimetric analysis was made under reduced nitrogen atmosphere from 0-300˚C using temperature gradients of 5˚C/min. Scans were routinely recorded as duplicate using DSC (v.23.10 .build 79), TA instruments, USA.
Thermo gravimetric analysis (TGA)
Approximately 5µg sample was loaded in the platinum TGA pan. Thermo gravimetric analysis made under reduced nitrogen atmosphere from 0-800˚C using temperature gradient of 10˚C/min. Scans were recorded as duplicate.
Gas chromatography-mass spectroscopy (GC-MS)
GC-MS analysis was performed in Shimadzu Model QP2010 with head space analyzer, quadrapole detector using DB-5 MS column (30m length X 0.25mm ID X 0.25μm) operated under following conditions viz., manual injection; initial oven temperature as 40˚C; injector temperature as 200˚C; Ion source temperature as 200˚C; interface temperature as 240˚C. Helium (99.999%) was used as a carrier gas at the flow rate of 1.36ml/min. Eluent from GC column was directly introduced into the source of mass spectra and spectra obtained compared with that of peaks with Wiley and NIST libraries and with published data.
1H NMR spectral analysis
1H NMR spectrum of the sample was recorded using JEOL ECA 500MHz spectrophotometer equipped with Delta version; irix platform operating at 500MHz (1H) and 125(13C). Sample concentration was maintained as 0.5mM per ml in 0.6ml duterated methanol solution (Deuterated, Sigma, USA, 99.99%) in a 5mm OD wilmad percisison NMR tube. 1H chemical shifts of solvent (7.27ppm) used as a secondary reference and referred to TMS signal using usual relationships. Proton NMR (1H/pNMR) spectra was collected as 16K points over an x-sweep till 20ppm, spectral width using a 90˚ pulse with a relaxation delay of 5 seconds.
Application studies
Coloring of fabrics: Three different types of fabrics were taken for the coloring study namely natural fibre (cotton), protein fibres (wool and silk) and synthetic fibres (polyester and polypropylene) and all the fibres were scoured, bleached before subjected to coloring. The fabric was introduced in a bath containing the pigment at the room temperature. The temperature was then gradually raised to 60˚C and maintained for 1hr for fixation. The fabric samples were then air dried at 100˚C for 60sec. A 10% shade was provided for dyeing all the samples.
Color strength evaluation
The color uptake on the different types of fabric was assessed by measuring its color strength (K/S) value using UV–Vis spectrophotometer. Kubelka Monk equation (K/S) defines a relationship between spectral reflectance of sample and its absorption (K) and scattering (S) characteristics. The color strength of the fabric was measured at minimum reflectance value (λmax) as a function of wavelength. The color strength (K/S) value was calculated using the formula given below:
(1)
Where K is the coefficient of absorption, S is the coefficient of scattering; and R is the reflectance value of the fabric at λmax. The hue and chroma values were calculated using L*, a*, b*, hue (ho), chroma (C*) values as follows.
(2)
(3)
(4)
(5)
(6)
Fastness testing
All the fabric samples (cotton, wool and silk, polyester and polypropylene) were tested for color fastness to washing according to ISO 687-1979. Fastness to rubbing and perspiration were tested according to ISO 766-1988 and ISO 971-1956, respectively. Color fastness to light was tested according to ISO 2454-1985.
Molecular docking study
In order to assess the nature of interaction of Asperyellone with silk protein, molecular docking study was performed using Autodock 4.2 software. The chemical structure of the Ligand, Asperyellone was retrieved from Pubchem database with CID no: 101600052 and the structure of silk protein (2K3N) were downloaded from Protein data bank. These are generally considered as the preliminary step in docking protocol. The second step which is creating the PDBQT, Grid parameter file (gpf) and docking parameter file (dpf) was prepared. Molecular docking was done using Cygwin and the results were analysed.
Aspergillus species strain AN01 isolated from soil samples showed complete growth in the CPD agar plate as shown in Figure 1a and when stained with Lactophenol cotton blue (Figure 1b) confirmed the Aspergillus family. Figure 1c depicts the yellow pigment production under culture conditions followed in the present study and Figure 1d illustrates the yellow pigment extracted from the biomass using acetone. The physiochemical properties of the yellow pigment were shown in Table 1.

Figure 1 Aspergillus species strain AN01 chosen for the present study a) Macroscopic view on growth of species in plate b) Microscopic view (Lactophenol cotton blue staining of spores) c) Yellow mycelial balls produced by Aspergillus species strain AN01 during growth d) A pure yellow pigment extracted from Aspergillus species strain AN01.
S.No |
Properties |
Pigment |
1 |
Color |
Yellow |
2 |
Odour |
Fruity odour |
3 |
pH |
6.5 |
4 |
Solubility in water |
Insoluble |
4 |
Solubility in organic solvents |
Soluble in Acetone, Dimethylsulfoxide, Methanol, Hexane, Benzene, Chloroform, Dichloromethane, Tetrahydrofuran, Ethylacetate, Acetonitrile, Isooctane. |
5 |
Light sensitivity |
Sensitive to visible light |
6 |
Presence of any elemental constituents |
Nil |
7 |
Presence of any salts |
Nil |
8 |
Flame test |
Charred at high flame |
9 |
Molecular formula |
C20H22O |
10 |
Molecular Weight |
278g/mol |
Table 1 Physiochemical properties of the yellow pigment extracted from Aspergillus species strain AN01
Instrumental analysis
UV spectral analysis of the purified pigment showed two absorption peaks at 280nm and 420nm. Figure 2a indicates the presence of aromatic ring with extended conjugation.12,14 FT-IR spectrum reveals the presence of mono-substituted phenyl rings and conjugated double bonds near 700, 1010, 1560cm-1 as shown in Figure 2b.11 Differential scanning caloirometry study showed that the melting point of the compound as 196.74˚C. Thermogravimetric analysis showed 54% weight loss at 302˚C which infers the presence of thermo stable compound as shown in Figures 2c & 2d respectively. The spectral results corroborates well with the results of Yu et al.,11 Mass spectrum of the compound shows molecular ion peak (Figure 3a) at m/e 281, 57 (ethyl ketone group), 77.10 (mono-substituted phenyl ring), and 265.25 (methyl group attached to the carbon chain) as reported by Jefferson,12 and Rao et al.13 The NMR spectrum of the yellow pigment was shown in Figure 3b. From the three protons triplet observed at δ 1.25 it was observed that the methyl group must attach to carbonyl group. The singlet observed at δ 2.1 suggests that methyl group is attached to double bond. The olefinic protons were observed in the range of δ 6.3-6.7 and the aromatic protons in δ 7.1-7.5 (subset of Figure 3b). The NMR spectrum and the fragments correlate with the compound Asperyellone as reported by Yu et al.11 With reference to NMR spectrum and GC-MS analyses, the molecular formula and the structure of the compound was determined as C20H22O and the relative structure of the compound shown in Figure 3b.

Figure 2 Instrumental analysis of the yellow pigment isolated from Aspergillus species strain AN01. (a) UV-Visible spectral analysis of Asperyellone pigment. (b)FT-IR Spectral details Asperyellone pigment. (c) DSC analysis of Asperyellone pigment. (d) TGA of Asperyellone pigment.


Figure 3 (a) GC-MS analysis of Asperyellone pigment. (b) 1H NMR (including the inset) spectral analysis of Asperyellone pigment and structure elucidated.
Coloring of fabrics with Asperyellone
The color strength (K/S values) of the fabrics in the presence of Asperyellone, without addition of any chemicals given in Table 2 and its reflectance curve illustrated in Figure 4a. The observations suggested that the color uptake is comparatively higher for protein fibre (K/S=2.11) followed by synthetic polypropylene fibre (K/S=2.29) and cotton fibre (K/S=0.79). Based on the nature of the fibres, shift in λmax was observed. In case of silk and PP the λmax is 400nm and for cotton, wool and polyester is 420nm.
Samples |
Color strength (K/S) |
L* |
a* |
b* |
Hue (H) |
Chroma(C) |
Cotton |
0.79 |
87.71 |
-9.17 |
30.26 |
73.14 |
31.62 |
Silk |
1.01 (λmax =400 nm) |
83.23 |
-7.26 |
28.06 |
-75.62 |
28.98 |
Wool |
2.11 |
76.79 |
-7.15 |
30.501 |
-76.91 |
31.33 |
PET |
0.52 |
83.89 |
-5.84 |
17.12 |
-71.16 |
18.09 |
PP |
2.29 (λmax =400 nm) |
82.63 |
-8.5 |
30.51 |
-74.43 |
31.67 |
Table 2 Color strength values for different types of fabrics colored with a yellow pigment Asperyellone extracted from Aspergillus species strain AN01
To further, the effect of concentration of Asperyellone on the color depth of the silk fabric was studied at 100%, 75%, 50% and 25% concentration and the results were shown in Table 3. The results suggested that while increasing the concentration, the color strength of the fabric is also increases from 0.47 to 1.44 and the related constants L*, a*, b* values were given in Table 3. All the samples follow the same reflectance path as shown in Figure 4b and its fastness properties assessed were shown in Table 4. The results on fastness study indicated that except light fastness all the fastnesses are coming under the good scale reading.
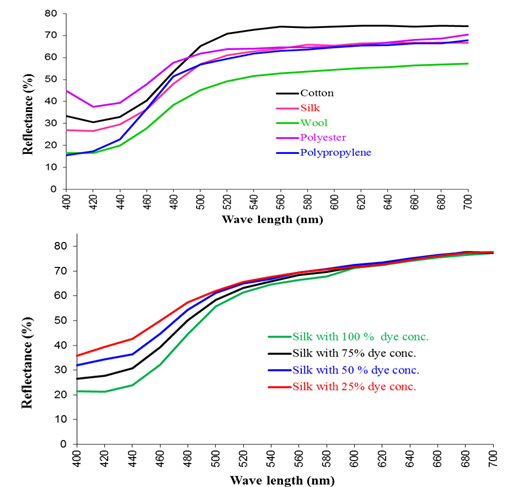
Figure 4 (a) Reflectance curve of Yellow pigment (Asperyellone) in different types of fabric. (b) Reflectance curve of silk fibre colored with different concentration of yellow pigment.
Silk |
Color strength (K/S) |
L* |
a* |
b* |
Hue (H) |
Chroma (C) |
100% |
1.44 |
84.37 |
-5.83 |
36.03 |
80.81 |
36.5 |
75% |
1.02 |
85.25 |
-4.93 |
29.08 |
80.38 |
29.49 |
50% |
0.72 |
85.94 |
-3.99 |
24 |
80.57 |
24.33 |
25% |
0.47 |
86.09 |
-3.14 |
18.79 |
80.51 |
19.05 |
Table 3 Color strength values for silk fabric at different concentrations of Asperyellone extracted from Aspergillus species strain AN01
Color fastness to |
Color change & staining |
||||
Cotton |
Silk |
Wool |
PET |
PP |
|
Washing |
3-4 |
4-5 |
4-5 |
3-4 |
4-5 |
Perspiration (Acid) |
3-4 |
4 |
4 |
4 |
4 |
Perspiration (Alkali) |
3-4 |
4 |
4 |
4 |
4 |
Rubbing Dry |
3-4 |
4 |
4 |
4 |
4 |
Rubbing Wet |
3-4 |
4 |
4 |
4 |
4 |
Light |
1 |
1 |
1 |
1 |
1 |
Table 4 Color fastness results for different types of fabrics colored with Asperyellone extracted from Aspergillus species strain AN01
The samples were tested for color fastness to washing and perspiration using Society of Dyers and Colorists (SDC) standard multifibre fabric. The standard grey scales for assessing change in color and staining properties were used to obtain ratings. Tables 4 & 5 outlines the results of the color fastness.
Color fastness to |
Color change & staining for silk |
|||
100% |
75% |
50% |
25% |
|
Washing |
4-5 |
4-5 |
4-5 |
4-5 |
Perspiration (Acid) |
4 |
4 |
4 |
4 |
Perspiration (Alkali) |
4 |
4 |
4 |
4 |
Rubbing Dry |
4 |
4 |
4 |
4 |
Wet |
4 |
4 |
4 |
4 |
Light |
1 |
1 |
1 |
1 |
Table 5 Color fastness results for silk fabric at various concentrations of Asperyellone extracted from Aspergillus species strain AN01
Tables 4 & 5 shows the color fastness results for various samples and indicate that all the samples have relatively good color fastness except cotton. The fastness to washing is good for all the fibres except polyester. Perspiration fastness for both acid and alkali received a grey scale rating of 4 for all the samples. But all the samples possess poor color fastness to light and received a blue scale rating of 1. Shirata et al.18 studied the dyeing property of violacein which was very sensitive to sunlight. In order to increase the resistance to light he subjected the dyed fabric to thiourea post treatment and observed that incorporation of thiourea imparts resistance to light. Figure 5 illustrates the optical image of the silk and wool after exposed to yellow pigment Asperyellone.

Figure 5 Fabrics Silk and wool colored with yellow pigment (Asperyellone) from Aspergillus species strain AN01.
All the observations with reference to coloring of fabrics especially silk fabric using microbial pigment suggested that the color exhaustion is at higher level in silk than wool, cotton and polymeric materials. Silk fabric coloring is a subject of interest and most of the dyes are sodium salts of organic acids and the better dye uptake is related with the pH, quality of water and duration. In the present study, since the yellow pigment is soluble in various organic solvents the exhaustion of the color is at appreciable level without generating any toxic wastes, asperyellone may be the suitable color pigment to improve the aesthetic appearance of the fabric. Further, the color uptake does not require any steaming to improve the exhaustion level. Furthermore, the reason for the high color exhaustion has been evaluated using molecular docking analysis.
Interaction of Asperyellone with silk protein peptides (Docking)
Docking was performed using Autodock 4.2 with Ligand CID no: 101600052 and the representative silk protein. Molecular docking study results in various ligand binding describes the best among them showed maximum interaction energy of around -5.4kcal/mol. It was observed that the Asperyellone showed surface binding interaction with the ser 130A residue of the silk protein as shown in Figure 6.
The yellow pigment Asperyellone extracted from Aspergillus species strain AN01 showed a significant application as a biocolorant for dyeing especially silk and wool. In silk, the effect of pigment concentration found to be directly proportional to color strength of the fabric. In color fastness study, all the fabric displayed a good scale reading of 4 & above except cotton. In all fabric, the color fades rapidly in the presence of sunlight because of its photosensitive nature. Studies have to be performed in order to decrease the fading nature of the fabric. Docking study revealed that Asperyellone showed surface binding interaction with Ser 130A residue of the silk protein.
There is no conflict of interest in terms of finance for this study.

©2019 Iswarya, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.