Journal of
eISSN: 2373-6453


Research Article Volume 3 Issue 3
1Department of Medical Lab Sciences (DMLS), Faculty of Health and Allied Sciences, Imperial College of Business Studies, Pakistan
2Atta-ur-Rehman School of Applied Biosciences, National University of Sciences and Technology, Pakistan
3Imperial Post Graduate Medical Institute (IPGMI), Faculty of Health and Allied Sciences, Imperial College of Business Studies, Pakistan
Correspondence: Ahmed Bilal Waqar, Associate Professor/Director, Imperial Post Graduate Medical Institute, Faculty of Health and Allied Sciences, Imperial College of Business Studies, Lahore, Pakistan, Tel (92) 3349686443
Received: April 26, 2016 | Published: May 12, 2016
Citation: Atif M, Raheel U, Alam F, Arshad HU, Baloch FUH, et al. (2016) Serotyping of Dengue Virus from Deadly Outbreaks of Pakistan. J Hum VirolRetrovirol 3(3): 00092. DOI: 10.15406/jhvrv.2016.03.00092
Dengue Dengue virus (DENV) belonging to the family Flaviviridae consists of 4 serotypes (DEN-1, 2, 3 & 4) causing severe illnesses like dengue fever (DF), Dengue Hemorrhagic Fever (DHF) and Dengue Shock Syndrome (DSS). Dengue fever has become an emerging disease in Pakistan during the past 5-7 years with an increase in the number of cases every year since 2006. The current study aimed at identifying the serotypes of dengue virus involved in 2011 outbreak of Pakistan. Samples were collected from all provinces of Pakistan. Patients were confirmed in the hospital by antibody captured Enzyme linked immunosorbent assay (ELISA). These ELISA positive serum samples were inoculated on HeLa cell line to raise the viral titer of dengue virus and then RT-PCR was done for serotyping of dengue virus. Initially D1, D2 primers were tried for serotyping of dengue virus but poor results were obtained.
Then serotype specific primers were designed having equal fragment length targeting NS1 gene. This approach was successful in serotyping of dengue virus from Pakistani population. On Real time RT-PCR, Dengue virus (DENV) was detected in 43.33% (n=26) patients out of 60 analyzed samples. Among 26 positive cases, 76.9% (n=20) were children and 23.1% (n=6) were adults and 46.1% (n=12) were male and 53.8 % (n=14) were females. ELISA results of 26 PCR positive cases show that 53.8% (n=14) were positive for IgM, 15.4 % (n=4) were positive for IgG and 30.8 % (n=8) were positive for both IgG and IgM. In this study DEN-2 was detected in 13.3% (n=8) patients, DEN-3 was detected in 16.67% (n=10) patients and DEN-2 & DEN-3 coinfection was detected in 13.3% (n=8) patients. We did not detected DEN-1 & DEN-4 in 2011 outbreak of Pakistan in 60 analyzed samples. By this study we can conclude that DEN-2 and DEN-3 were the main serotypes involved in 2011 outbreaks in Pakistan.
Keywords:Dengue, DHF, Dengue virus, RNA, Dengue fever, DF, Serotyping, DSS, Dengue hemorrhagic fever, Viral infection
DHF, Dengue Hemorrhagic Fever; DF, Dengue Fever; ELISA, Enzyme Linked Immunosorbent Assay; DENV, Dengue Virus; YFV, Yellow Fever Virus; JEV, Japanese Encephalitis Virus; WNV, West Nile Virus; MVEV, Murray Valley Encephalitis Virus; IRB, Institutional Review Board; ASAB, Atta-ur-Rahman School of Applied Biosciences; MEM, Minimum Essential Media; PMNs, polymorph Nuclear Leukocytes; DSS, Dengue Shock Syndrome
Dengue virus (DENV) has emerged as the most important mosquito born viral infection of 21st century.1 DENV belongs to the genus flavivirus which contains many other lethal human viruses including Yellow fever virus (YFV), Japanese encephalitis virus (JEV), West Nile virus (WNV) and Murray Valley encephalitis virus (MVEV).2-4 DENV infection causes a broad spectrum of sign & symptoms which varies from self-limiting DF to life threatening DHF and DSS.5-6
DENV is a positive sense RNA virus having a genome size of 11Kb including 3 structural & 7 nonstructural genes.7-8 DENV has four predominant serotypes DEN1 to DEN4 based on differences in antigenic domains.7 DENV spread to the humans by the bite of infected Aedes aegypti and Aedes albopictus mosquitos.9 DENV infection is endemic in approximately 100 countries of the world and it is the leading cause of mosquito borne viral infection of humans. In many Asian countries, DENV is the main reason of hospitalization and death.10
In the last 30 years, the number of countries reporting dengue fever outbreaks has increased by 10-fold including countries of Pacific-Asian region, the Americas, the Middle East, and Africa. According to WHO, 2.5 billion people around the globe are at risk of dengue fever encompassing around 100 countries with 50 million yearly cases, out of which 5,00,000 proceed into DHF. The Annual deaths due to dengue infection are from 20,000 to 25,000 and majority of them are children.11,12
DENV is prevalent in parts of Asia, Central and South America and Africa.13 During the last decade, epidemics of DHF have occurred in Pakistan, India, Bangladesh, China, Sri Lanka, and Maldives. All four serotypes of dengue cause similar type of infection, but DENV-2 and DENV-3 are more often associated with severe and fatal DHF. DENV-2 and DENV-3 are the most frequently found genotypes in South East Asia.14,15 In Pakistan DF has caused many outbreaks from 1994 to 2011.16 DF was first reported from Pakistan in 1982 in which out of 174 patients 12 patients were victims of DF.17
The first major outbreak of DHF was documented in 1994 from Pakistan. In this outbreak 15 out of 16 patients were positive for dengue virus IgM antibodies and DEN-1 and DEN-2 serotypes were reported from this outbreak.18 In 2005 & 2006 two outbreaks of DHF were reported from Karachi, Pakistan. DEN-3 was isolated from 2005 outbreak while in 2006 outbreak co-infection of DEN-2 and DEN-3 was found.19,20 In 2008, another outbreak of dengue fever was observed in Lahore where a large number of people were infected with DENV. Co-infection of DEN-2, DEN-3 and DEN-4 was found in this outbreak.21 DENV circulates in Pakistan throughout the year with a peak incidence in the months of July to October (post monsoon season). This period is further endangered by the floods. Still there is no treatment or vaccine strategy available to cure dengue fever. Early identification of the dengue virus remains a keystone for the treatment of dengue fever.22
Sample Collection
This study was approved by Institutional Review Board (IRB), ASAB, NUST. DENV positive samples were collected from various locations of the country. 131 Serum and blood samples were collected from Meo Hospital Lahore, Jinnah Hospital Lahore, Children’s Hospital Lahore, Holy Family Hospital Rawalpindi and PIMS Islamabad. All samples were put into ice after collection and were transported to Atta-ur-Rahman School of Applied Biosciences (ASAB), NUST and stored at -150oC until analysis.
Dengue virus culturing on HeLa cell line
HeLa cells line was used for propagation of DENV. Virus was grown at 37˚C for three days and then harvested. Minimum Essential Media (MEM) was used for maintenance of HeLa cells which contains a balanced salt solution together with 5% fetal calf serum (FCS), 1% sodium bicarbonate, 2% L glutamine, 2% penicillin, amphotericin B, streptomycin and 1% non-essential amino acid. Twenty-five sq cm screw-capped flat bottomed flasks were seeded with 3 mL of cells having got 70-80% confluency. 140 µL of patient’s serum was passed through a 0.2 µm to remove any microbial contaminants and then inoculated into each flask with confluent mono layer of HeLa cells. 140µL of PBS (pH-7.4) was also added in a separate flask as a control. The inoculated flask was incubated for 45 minutes at 37˚C with agitation in shaking water bath incubator (memmert water baths WNB7-45). Maintenance media (growth media with 10% FCS and 0.5 % ampicillin) in a quantity of 7ml was then added to the flask and flask was incubated at 37°C maintained for three days. Cells were regularly observed under inverted microscope (LabMed TM400) for the detection of visible cytopathic effect (CPE).
Imaging of the infected cell line
HeLa cell line infected with DENV was observed under inverted microscope (LabMed TM400) and images were captured with inbuilt camera. Images were taken at an interval of 24 hours, for three days. Finally imaging was performed on day 3 post infection and images were saved.
NS1 primers used for serotyping of DENV
Initially for serotyping of DENV D1, D2 primers were used but unsatisfactory results were obtained. In an attempt to serotype Dengue virus from Pakistan population another approach was used in this study using NS1 primers for all four serotypes of dengue virus. We synthesized NS1 primers for all four serotypes of dengue virus and then 60 samples were run separately with D-1, D-2, D-3 and D-4 primers. This approach was successful in serotyping of dengue virus. Sequences of NS1 primers used for serotyping are given in Table 1.
Sr.# |
Serotype |
Forward |
Reverse |
1 |
DENV-1 |
5َ GGA TCC AGG GGA AGT GGA CAG CTT TTC3َ |
5َ AAG CTT TGG TTA ATG GTT TGC ATC CAA GAG A3َ |
2 |
DENV-2 |
5َAACTCGAGAAAGAATGGATAGTGGTTGCGTTGTGAG3َ |
5َAAGGTACCTTAGGCTGTGACCAAGGAGTTAC3َ |
3 |
DENV-3 |
5َGGATCCATTACACTCTATCTGGGAG3َ |
5َAAGCTTGTTGTCCATCTTTCCAC3َ |
4 |
DENV-4 |
5َGGA TCC ACA TGG GTT GTG TGG TGT CAT3َ |
5َAAG CTT GCC GTC ACC TGT GAT TTA AC3 |
Table 1 Showing sequence of primers used for serotyping of DENV
RNA extraction
RNA extraction was done using Qaigen RNA extraction kit according to manufacturer’s protocol. Extracted RNA was quantified on BioPhotometer plus (Eppendorf Germany).
Reverse transcription
After RNA extraction cDNA was synthesis immediately by Reverse Transcription (RT). For cDNA synthesis, 17.5µL RNA 80 ng/µL, 2.5µL NS1-DENVR3 specific reverse Oligonucleotides (50 picomole), 1µL Dithiothreitol (DTT) 2.5µM and 4µL DEPC water were initially incubated at 70oC for 10 minutes. RT reaction was carried out with 10µL of 5X RT buffer, 2.5 µL of 10mMdNTPs 500µM (Fermentas, USA), 1µL of 40U of RNAse inhibitor (Fermentas, USA), 2 µL of 200U MuLV RT enzyme (Fermentas, USA), 9.5µL of DEPC water in a final volume of 50µL. During pipetting reaction mixture was placed on ice and after a short spin it is placed in ESCO Thermal Cycler system (Swift MaxPro Singapore). RT reaction was done at 42oC for 60 minutes. cDNA was then immediately processed for PCR amplification or otherwise stored at -20oCprior to PCR amplification.
Polymerase chain reaction (PCR)
PCR reaction was performed with 7µL of RT-cDNA, 2mM dNTPs 2µL, 50 pmol forward NS1-DENVF 1µL and NS1-DENVR reverse 1µL oligonucleotides, Taq polymerase 1µL (Fermentas, USA), 10X Taq buffer 2µL (Fermentas, USA), 25Mm MgCl2 1µL, Nuclease free (NF) water 5µL (Fermentas, USA) to a final volume of 20µL. During pipetting reaction mixture was placed on ice and was spin before addition of Taq polymerase (Table 2). After addition of Taq polymerase reaction mixture was immediately placed in ESCO Thermal Cycler system and run for 35 cycles at the following profile. 94oC: 4 minutes, 94oC: 45 seconds, 55oC: 1 minute, 72oC: 2, 72oC: 10 minutes.
Sr.# |
Parameter |
Total |
Percent |
1 |
DENV II Only |
8 |
13.30% |
2 |
DENV III Only |
10 |
16.70% |
3 |
DENV II & DENV III |
8 |
13.30% |
4 |
IgM Only |
14 |
53.80% |
5 |
IgG Only |
4 |
15.40% |
6 |
IgM & IgG |
8 |
30.80% |
Table 1 Showing percentage of positive cases of ELISA and PCR
Agarose gel electrophoresis
PCR product was run on 1 % TBE agarose gel, prepared by adding 0.4 g agarose in 40mL of 1X TBE (Tris borate Ethylenediamine tetra acetic acid) (10X TBE (Tris Base: 108g, 0.5M EDTA (pH:8.3): 20ML, Boric Acid: 55g dissolved in 1000 ML, Ph adjusted to 8.3 and diluted to 1X) and boiled for 1 min in microwave oven. Upon cooling to 55-60˚C, 4 µL of Ethidium bromide was added (Table 3). The gel was allowed to cool to 50°C and then poured in the gel caster (Wealtec (USA) electrophoresis apparatus) for solidification. TBE buffer was used for electrophoresis. The DNA samples of 7µL were prepared by adding 6x loading dye (5:1 ratio). Samples and DNA ladder was loaded and run at a voltage of 80V. To visualize the bands gel documentation system (Wealtec, Dolphin-Doc, USA) was used and pictures were saved.
Sr.# |
Parameter |
Total |
Platelet count less than 100,000/ml |
Percent |
1 |
DENV II Only |
8 |
4 |
50% |
2 |
DENV III Only |
10 |
10 |
100% |
3 |
DENV II & DENV III |
8 |
8 |
100% |
4 |
IgM Only |
14 |
12 |
86% |
5 |
IgG Only |
4 |
4 |
100% |
6 |
IgM & IgG |
8 |
6 |
75% |
Table 3 Showing percentage of positive cases having low platelet count
HeLa cell line infection with serum of dengue virus patients from Pakistani population
It is very difficult to isolate dengue virus directly from serum samples due to several factors including sample storage, transportation, positive sense RNA genome, easily degraded viral RNA. Therefore HeLa cell line was infected with serum of DENV positive patients for DENV propagation and PBS as positive control. Cell line was observed before infection and post-infection for cytopathic effects (CPE) for three days. Images of the infected cell line were obtained after 24 hours intervals. Cell viability was more than 80% before inoculation of DENV3 positive serum but cell death reduced the cell viability to less than 10%, confirming that DENV propagated on HeLa cells leading to massive cell death. Cytopathic effects were visible after 24 hour; infected HeLa cells were exhibiting increasing signs of cell death with the passage of time. Imaging of infected cell line showed clumps of dead cells floating in the supernatant, while few living cells were still attached to the surface of flask. Imaging of infected and uninfected HeLa cell line after day 1, 2 and 3 are shown in (Figure 1).
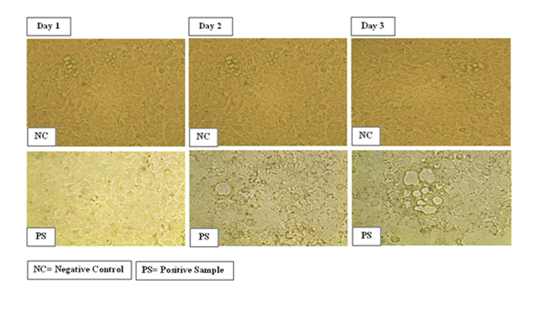
Figure 1 HeLa infected cell line with negative control and DENV infected serum after day1, 2 and 3. (Pictures taken at 20X).
Serotyping of dengue virus
Out of 131 collected samples, 60 samples were processed for serotyping of DENV through RT-PCR. RNA was extracted using Qaigen RNA extraction kit; cDNA was prepared using reverse transcription (RT), the DNA product was run for serotyping by using PCR targeting NS1 gene. The serotyping results were analyzed on 1% agarose gel as shown in (Figure 2). Serotyping was done by using serotype specific primers having same fragment size. Positive cases were divided into 3 groups based on the results of serotyping (Group 1= DEN-2 positive cases, group 2 = DEN-3 positive cases, group 3 = DEN-2 & DEN -3 coinfection). Results of Serotyping and collected data was entered and analyzed in SPSS 19. Results were displayed in the form of graphs. Out of 60 analyzed samples, 8 samples were positive for DEN-2 (13.3 %), 10 for DEN-3(16.7%), 8 samples were positive for DEN-2 & DEN-3 (13.3%) coinfection and 34 samples (56.7%) were negative for DENV infection (Figure 3). Out of 26 positive cases 20 (76.9%) were children and 6 (23.1) were adults (Figure 4). Out of 26 positive cases 12 (46.1 %) were male and 14 (53.8 %) were females (Figure 4). Out of 26 positive cases 14 (53.8) were positive for IgM, 4 (15.4 %) were positive for IgG and 8 (30.8 %) were positive for both IgG and IgM (Figure 4). In group 1 All 8 cases in group 1 were children. Out of 8 positive cases in group 1, 4(50%) were male and 4(50 %) were females. Out of 10 positive cases in group 2, 8(80%) were children and 2(20%) was adult. Out of 10 positive cases in group 2, 2(20%) was male and 8(80%) were females. Out of 8 positive cases in group 2, 4(50%) were children and 4(50%) were adults. Out of 8 positive cases in group 3, 6(75%) were male and 3(25%) was female.
Dengue virus is the most destructive arboviral pathogen affecting more than 100 countries of the world.11 It is a serious health issue in South East Asia including Pakistan with many outbreaks in the last decade. Due to poor health facilities, lack of vector control, expensive diagnosis and no licensed vaccine the number of dengue virus cases is increasing every year in Pakistan.22 Accurate and timely diagnosis of dengue virus is essential in the management of the disease.
For successful isolation of dengue virus from serum samples, dengue virus was cultivated on HeLa cell line. Previous studies show that DENV can be isolated from various cells like polymorph nuclear leukocytes (PMNs), attached phagocytic monocytes, macrophages and also some non-adherent leukocytes.23,24 Dengue virus in vitro can infect different primary lineages and establish cell lines like endothelial and fibroblast cells, myeloid-derived cells, and lymphocytes.25 DENV can infect and propagate in various human and insect cell lines like C6/36, HeLa and THP-1.26 Dengue virus can grow on various mammalian and insect cell lines like BHK-21, C6/36, HeLa, HepG2 and THP-1.27 The most widely employed cell line for the propagation of DENV is C6/36, which is isolated from A. albopictus.28 Two other most common insect cell lines allowing efficient DENV growth are AP-61 from A. pseudoscutellaris and TRA-284 from T amboinensis. TRA-284-SF cell line can adapt to serum free media so it is cost effective as compared to other two cell lines.29 Amongst human cell lines when infected with DENV maximum viral yield was observed in A549 (lung carcinoma cell line) and HepG2 (a hepatoma cell line).26 HeLa is a cancer cell line and is widely used for propagation of different viruses. HeLa cells have the property of adherence and the mostly grow sticking at the bottom of cell culture flask; HeLa was the first cell line to grow outside the human body. 70-80% confluent cells in the cell culture flask are split for further passages. Cells can be passage up to 20 times and they grow best at 37˚C inside a carbon dioxide CO2 incubator. In the present study HeLa cell line was chosen because of its rapid growth rate and easy maintenance under laboratory conditions.
In the present study 60 samples out of 131 collected samples were run for serotyping by the most sensitive real-time PCR technique. Initially we used D1 and D2 primers for serotyping of Dengue viruses but Poor results were obtained from these primers because already published primers for serotyping of dengue virus from different countries at that time did not worked well with Pakistani dengue virus serotypes. To overcome this problem we specially designed primers specific for NS1 region of all four serotypes of Dengue viruses reported from Pakistan. Studies have shown that primers specific for Envelop and NS1 region have also been used in the past.30 In the area where our study is being conducted no serotyping is being done commercially of dengue viruses. So the present technique used in the study can also be used commercially for serotyping of the dengue viruses.
All four serotypes (DEN-1, 2, 3 &4) of DENV cause DF but DEN-2 and DEN-3 are the most prevalent DENV serotypes worldwide. In the current study also DEN-2 and DEN-3 were the predominant serotypes found circulating in 2011 outbreak of Pakistan which is consistent with the previous studies. Out of 26 positive cases, 8 samples were positive for DEN-2, 10 with DEN-3 and 8 had coinfection with DEN-2 & DEN-3. Fatima et al.18 also reported the similar findings in 2007, 2008 and 2009 outbreak of Pakistan with gradually increase in appearance of DEN-3 in latter outbreaks.31 In 2007 outbreak DEN-2 was predominant with less number of DEN-3 while in outbreaks of 2008 and 2009 DEN-3 was present in large numbers along with DEN-2. Humayoun et al.20 also reported DEN-3 in 2008 outbreak of Lahore.20
DEN-3 was first reported by Jamil et al.18 in 2005 outbreak of Karachi.18 From then DEN-3 along with DEN-2 has become a major cause of DHF/DSS in the country. Other serotypes of DENV (DEN-1 and DEN-4) are rarely reported from Pakistan. Related researches from India also document the prevalence of DEN-2 & DEN-3 with increased incidence of DHF and DSS.
DENV easily undergo genetic mutations because it replicates by RNA polymerase which does not possess a proof reading mechanism. These mutations lead to the emergence of new serotypes and genotypes of DENV. South East Asia is considered to be the hub of these genetic mutations as new strains of DENV mostly emerge from this region including DEN-3. DENV serotype 3 responsible for 2005 outbreak of Karachi was found to be very similar to the Indian DEN-3.18,19 Fatima et al.31 found that the DEN-3 of Pakistan had 99% homology with Sri Lankan DEN-3 strain.31
In the current study more children (20) were found positive for dengue virus than adults (6) which is consistent with the previous studies.32-34 reporting the prevalence of DF and DHF in childhood, especially in South East Asia where mean age of cases were reported to be under fifteen.22 Günther et al.29 also reported the predominance of DF and DHF in children with 11-15-year-old group being the most affected.35 Knowledge of the age-specific risk of the dengue is critical for rational implementation of dengue vaccines and anti-viral drugs to pre-school–aged children to achieve greatest impact of disease burden and mortality.
No difference in the positive results among the gender was observed. Out of 26 positive cases, 12 were males and 14 were females. This data is consistent with the previous reports suggesting no difference, a male excess, or a female excess in dengue patient populations.36-40 Bhaskar et al.30 also found no difference in male to female ratio in an observational study in Chennai, south India.41 Out of the 26 positive cases in this study, 14 were positive for IgM, 4 for IgG and 8 were positive for both IgM & IgG, which shows that most samples in this study were of primary dengue virus infection.
Dengue virus positive cases were divided into 3 groups based on the serotypes of dengue virus (Group 1= DEN-2 positive cases, group 2 = DEN-3 positive cases, group 3 = DEN-2 & DEN -3 coinfection). No significant results were found in group 1. In group 2, 4:1 ratio of child versus adults was observed which again show that DEN-3 is more common in children. Among gender, male to female ratio of 1:4 was observed which is not consistent with the previous reports documenting no gender biasness in dengue cases. This deviation is due to small sample size. In group 3 we found 3:1 male to female ratio which is again a limitation of the study due to small sample size. No other significant difference results were found in group 3. Serotyping strategy used in this study can be used for research and commercial purposes as it gives better results than the conventional strategy used with round 1 and 2 primers. No lab in Pakistan offer serotyping facility so our approach can be used for serotyping of dengue virus from clinical samples.
None.
None.

©2016 Atif, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.