Journal of
eISSN: 2373-6453


Literature Review Volume 11 Issue 1
Hakim’s Lab, Department of Biology, School of STEM, Diné College, AZ, USA
Correspondence: Shazia Tabassum Hakim, Hakim’s Lab, Department of Microbiology/Biomedical Sciences, School of STEM, Diné College, Tuba City, AZ, 86045, USA, Tel (928)-283-5113 Ext. (O) 7520; Ext (Research Lab, 7538)
Received: July 28, 2024 | Published: August 8, 2024
Citation: Cayatineto HW, Hakim ST. Prospective approaches to target bovine viral diarrhea virus and hepatitis C virus using miRNA-based inhibitors. J Hum Virol Retrovirol. 2024;11(1):36-47. DOI: 10.15406/jhvrv.2024.11.00278
Flaviviruses are a family of positive - single stranded RNA viruses, which includes Yellow Fever viruses (YFV), Dengue viruses (DENV), Japanese encephalitis (JEV), West Nile viruses (WNV), Zika viruses (ZIKV), Bovine Viral Diarrhea virus (BVDV), and Hepatitis C virus (HCV or Hepatitis C). Majority of these viruses are mostly carried by mosquitoes and are transmitted through mosquito bites or through contaminated blood or other blood products. As of now, there are vaccines available for most of these viruses, but some are still in development and research. HCV is one of the leading cause of liver cirrhosis, chronic hepatitis C, and liver cancers when left untreated.Currently, there is no vaccine available for this virus. That is why, HCV remains a threat for public health. Due to genomic similarities between HCV and Bovine Viral Diarrhea Virus (BVDV), BVDV is widely used as a surrogate model in studies related to HCV and its therapeutics. Hence, identifying a suitable target miRNA that could bind to the nucleocapsid protein gene of BVDV to inhibit viral replication is the main objective of this study and maybe later the same miRNA can be used for inhibition of HCV. The aim of this review is to highlight the importance of miRNAs targets, the impacts of Hepatitis C, and how miRNAs are being utilized as antivirals and vaccines.
Keywords: Flavivirus, Hepatitis C Viruses, Bovine Viral Diarrhea Virus, microRNA, Antiviral Therapy, RNA viruses.
YFV, yellow fever viruses; DENV, dengue viruses; JEV, Japanese encephalitis; WNV, West Nile viruses; ZIKV, Zika viruses; BVDV, bovine viral diarrhea virus; HCV or hepatitis C, hepatitis C virus
Virology of flaviviruses
The Flaviviruses are members of the family Flaviviridae and are enveloped viruses with a single stranded, positive sense RNA viruses,1 which is divided into three genres: flavivirus, pestivirus, and hepaciviruses2 and comprises over 70 viruses, which includes dengue virus (DENV) viruses, Japanese encephalitis (JE) viruses, St. Louis encephalitis (SLEV) virus, yellow fever (YFV) virus, and tick-borne encephalitis virus which are all important human pathogens.3–6 Flaviviruses also include members of West Nile (WNV) virus, Zika virus, and Hepatitis C viruses (HCV), which are among the hepatoviruses, and also belong to the flaviviruses family. This family even includes other important animal pathogens, such as Bovine Viral Diarrhea virus (BVDV).7 Flaviviruses share a common virion structure and among these viruses DENV and ZIKA are well characterized in this family.8 During infection, the genome is translated into a single polyprotein, which is then processed into the structural and nonstructural proteins (NS).9 The genome of the Flaviviridae family is organized as 5’-CAP(Type-I)-5’UTR-C-prim-E–NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-3’UTR10 and just one open reading frame (ORF) flanked by untranslated 5’ and 3’ regions (UTRs).11 In a study done by Ng, C, W., et al, the non-coding 5’UTR of flaviviruses comprise about 100 nucleotides in length, as the 3’UTR ranges from 400 to 700 nucleotides in length, depending on the virus (2017).
Virology of bovine viral diarrhea virus
Pestiviruses genus includes 4 species: BVDV 1 and 2, classical swine fever virus (CSFV) and border disease virus (BDV),12–16 which are a group of important animal pathogens that affect cattle, pigs, and sheep (Xu J, et al 1997). There are 2 subtypes of BVDV: BVDV type 1 and type 2, according to Jackov et al, BVDV Type 1 is a causative agent of bovine viral diarrhea and mucosal disease, while type 2 isolates may cause hemorrhagic syndrome with high mortality rate among cattle (2008) (Figure 1).
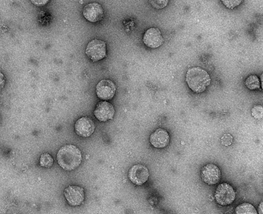
Figure 1 Electron microscopy of Bovine Viral Diarrhea Virus virions.17
BVDV virus particles range between 40 – 60 nm in diameter,18 and the genome is 12.3 kilo-bases in size.19 The viral proteins of BVDV are organized in the following order in the polyprotein: NH2-Npro-C-Erns-E1-E2-p7-NS2- NS3-NS4A-NS4B-NS5A-NS5B-COOH,20–23 which is quite similar to that of HCV Polyprotein. All viral structural proteins consist of autoprotease Npro, capsid protein C and the glycoproteins Erns, E1 and the E2,24,25 while the nonstructural (NS) proteins consist of the p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B.7,23 For viral attachment, BVDV attaches to the CD46bov26 and utilizing the glycoproteins E1 and E2; gp53 and gp48 receptors.27 Where, Omari, E, K., et al,28 clarifies the role of pestivirus glycoprotein E2 in viral fusion with host cells (2013) (Figure 2).28
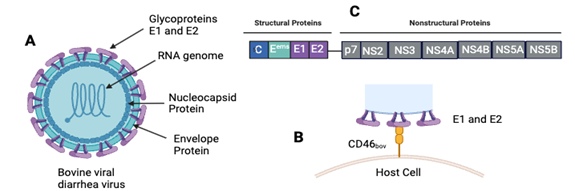
Figure 2 A. Virus Structure of BVDV. B. Viral Attachment of the E1 and E2 proteins to the CD46bov receptors in the host cell. C. Polyprotein organization of the structural protein and nonstructural Proteins.
BVDV transmission
Possible transmission of BVDV among cattle includes fomites, such as feed, water, and equipment such as the nose tongues, milk bottle nipples, needles, palpitations, secretions and excretion of urine feces, mucus, milk, and other contaminated minerals.29 When cattle are exposed, they usually recover and shed the virus temporarily, however pregnant cattle are more susceptible and the outcome depends on the gestational stage of the fetus (Fulton, R W et al., (2000).
Pathogenesis
Acute and persistent BVDV infections of pregnant cows are often accompanied by BVDV virus transmission into the fetuses in which the infections may result in abortions, teratogenic changes or delivery of persistently infected, immunotolerant calves, depending on the gestation.30 While cattle are the main host, BVDV infects various cattle including Bisons and can cause immune dysfunction and infection which result in asymptomatic infections and seroconversion or a variety of pathologies including fatal mucosal disease (Apapov, E. et al., 2003). If a cow is pregnant, the fetus will eventually become infected.31 The virus has the ability to cause transplacental infection resulting indifferent outcomes depending on the stage which would lead to fetal death, malformation, acute syndromes of the neonate, immune tolerance and lifelong viral persistence.32 Disease associated with BVDV can range from clinically inappropriate to severe, even with the availability of vaccines.33
Virology of hepatitis C virus
Like all members of the Flaviviruses, HCV contains a (+) stranded RNA genome, which contains a 9.6-kb with one long open reading frame (ORF) encoding a polyprotein that is co- and post translationally cleaved into structural proteins, that is a single stranded RNA of the positive polarity consists of a long ORF and 5’ and 3’ non-translated regions (NTRS).7,34 The HCV particles are enveloped with a diameter of 55 – 65 nm.35 The genome polyprotein organization of HCV are arranged in the following order: NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH,36–40 where the Core (C) and the E1, E2 are the major structural proteins that comprises the virions (virus particles),41 while the nonstructural proteins (NS) consists of the p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B.42–46 HCV, as of now, is one of the major concerns to public health, due to the lack of a suitable cell culture system to retain and maintain growth of the virus. Due to this drawback, there is no vaccine have been developed so far to prevent the disease. This is the reason why members of the Pestivirus groups of the Flaviviridae family that are closely related to HCV are widely used as a surrogate model for HCV (Figure 3) (Figure 4).47,48
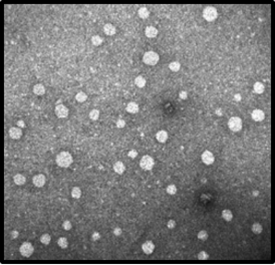
Figure 3 Electron microscopy of Hepatitis C Virus virions.48
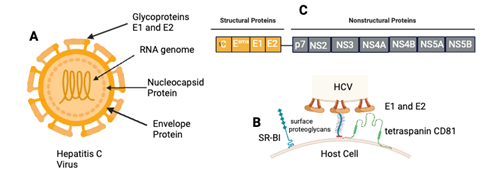
Figure 4 Overview HCV virus structure. A. The viral protein structure of HCV. B. The receptors of host cells for HCV entry. C. Polyprotein genome arrangement.
HCV transmission
HCV is a blood borne-virus, but there are other major routes of transmission, which includes sharing injection needles, drugs-abuse, which accounts for most infections, blood contaminated needles, transfusion of contaminated blood, and blood products, and unsafe/traumatic sexual practices.49–52 In a study done by Nijemijer MB et al.,53 they validated that there is evidence that acute HCV infections have occurred among MSM (men who have sex with men) who are HIV- positive and have become a risk factor. Nijemijer MB et al.,53 also states that HCV infection among MSM has increased from 0.07/100 person a year in 1990 to 1.8 per 100 person a year in the year of 2014 (2019). There were also studies indicated that in the early 1990s, a study showed an increased prevalence of HCV antibodies in alcoholic patients, with up to 30%-40% prevalence of chronic HCV infection reported in this population54 and in patients with cirrhosis had a greater total lifetime alcohol consumption.55
HCV pathogenesis
Acute HCV infection leads to more than 70% of patients to the development of chronic hepatitis and then cirrhosis and hepatocellular carcinoma56 despite antiviral therapeutics.57 Cirrhosis, portal hypertension, hepatic decompensation, and hepatocellular carcinoma have been reported due to chronic HCV infection.58 When left untreated, HCV can lead into Chronic HCV (CHC) infection, which progresses rather slowly after infection for most individuals and patients are often asymptomatic until they develop liver disease resulting in delayed diagnosis.59
Epidemiology of HCV
Worldwide, approximately 200 million people are infected with HCV and at serious risk of developing chronic hepatitis and hepatocellular carcinoma, no vaccines are available, and current therapies fail to eliminate the virus from a large number of patients.7 Approximately 2-4 million new infections occurring each year60 and over 350000 people die each year from hepatitis C related liver diseases.51,58,61 Furthermore, HCV is a leading factor of liver transplantation in the United States, and this has caused major implications in the present era of organ shortages.62 Moosay HS et al.,58 also mentions that more than 50% of hepatocellular carcinoma cases in the endemic population have happened due to chronic HCV infection and consisted of more than 6% of cirrhosis around the world (2017). In addition, there are problems in different countries; China, India, and the United States, which are the three most populous countries around the world, and they are also the top three countries for the burden of disease associated with HCV infections.63 Yang J et al.63 goes on to say that it was estimated that 6.2 million new HCV infections, 0.54 million HCV related deaths reported in 2019, with an increase of 25.4, and 43.6 % from the year 1990 and the numbers are still changing as the years go by (2023).
HCV impacts on indigenous communities
In the United States American Indian/Alaskan Native (AI/AN) are disproportionately affected by HCV infections. In the U.S., AI/AN and Canadian Aboriginal peoples have a higher prevalence of liver disease than other peoples.64 and validates indicate that chronic liver disease is the 5th leading cause of mortality for AI/AN peoples.65,66 Rempel J., & Uhanova, J further indicates that in Colonized countries, the prevalence of HCV infection in indigenous populations tends to be higher than non-indigenous populations (2012).65 In 2019, the U.S. Department of health and Human Services Office of Minority Health reported 9.05 cases per 100,000 and 9.08 death rates per 100,000. Dena Smith (2020) states that in the year of 2019 alone, approximately 35,000 cases of acute HCV among American Indians/Alaskan Native were reported including 3,887 AI/AN who are living with CHC that was well above the 2019 target of 41,467 estimated HCV infection in the U.S. (Figure 5). Although antiviral agents show great efficacy in HCV treatment, it seems that the global burden of liver disease does not decrease (Sharifnia, Z et al., 2019).
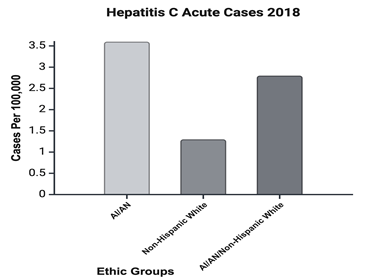
Figure 5 HCV cases per 100,000 among AI/AN compared to the cases among white communities. 2018 report by the Office of Minority Health.
Clinical virological diagnostics of hepatitis C
Viral serology and molecular assay for HCV have played roles in identifying the infection (Cloherty, G., et al., 2016) and is based on two types of laboratory test; serologic assays to detect anti-HCV antibodies, which is also known as the indirect test and using EIA or enzyme linked immunosorbent assay (ELISA) (Gupta, E., Baipai, M., & Choudhary, A., 2014).67 Guptae et al, and other researchers agree that HCV can also be detected through quantification of the RNA genome, by utilizing polymerase chain reaction (PCR) and/or real-time PCR (2014).
HCV cell culture system
Unveiling a robust cell culture system for HCV represents a valuable tool for in vitro studies and is still hampering screening of antiviral strategies,68 however without a known system this remains as a major obstacle for vaccine development. The main reason is that the infectious genome of HCV upon infection, failed to replicate in cell cultures. But engineering of HCV replicons to express a drug-selectable gene made it possible for the RNA replication in cell cultures.69 In 1992 HCV was successfully grown in human T-Lymphocyte MOLT-4Ma and HPB-Ma cell lines that were pre infected with murine retroviruses.70 Durverlie, & Wychwoski, mentions that other cell lines such as human hepatoma cell lines or Huh 7 and Hep-G2 could support the HCV replication, however these are not reliable and could not be used for studying viral cycle and screening for antiviral drugs (2007).70
In vitro replicons using JFH1 based system
There are a few robust methods by which HCV virions were grown and studied in vitro. Development of HCV RNA replicons was a breakthrough; however, these replicons do not undergo a complete replication cycle, and antiviral compounds will not identify early targets.71 Despite the fact that there is no validated cell culture system available to support the full viral life cycle, there have been methods developed to use the JFH1 base system for HCV genotype 1, 2 and 3, but still no known systems for genotypes 4, 5, and 6.72 Scheel et al discusses that in their investigation, they were able to establish a cell culture system to study HCV, however this is only the first step to uncover a new method to maintain the full infectious cycle of Hepatitis C (2008).72
Hepatitis C infection cycle
The infection cycle of HCV is not fully understood, due to that lack of an in vitro model system,73 but, just like many viruses replications, HCV replication can be divided into several stages, viral attachment, viral entry, release of genome, protein translation, RNA replication, viral assembly, and release of new virus (Li, H., Yang, C. & Lo, S., 2021). Because the RNA can act as an mRNA, viral proteins are translated in the cytoplasm by an RNA dependent – RNA polymerase, rather than entering the nucleus. HCV RNA replication takes place within the replication organelles (RO) of the endoplasmic reticulum (ER) (Li, H., Tang, C., & Lo, S., 2021). The (+) stranded RNA carries a single long open reading frame that encodes a polyprotein of 3,010 amino acids and generates series of proteolytic cleavages that are activated by peptidase and two viral proteinases amidated by host cell signals.74 The virus particles contain the core, the envelope glycoproteins E1, and E2.75 E1 has up to 6 glycosylation sites and E2, which as 11 glycosylation sites, that are used to attach to the host cell (Charles, M, R., 2011). HCV attaches to the multiple surface proteins called the scavenger receptor class B type 1, the low-density lipoprotein receptor (LDLr), and to the tetraspanin Cluster of Differentiation 81 protein for viral entry76 on the hepatocytes of the liver. HCV also binds to an additional receptor known as the claudin-1 and occludin proteins. The virus is then taken up via endocytosis where the RNA is released into the cytoplasm and is translated into viral proteins that make up the structure of the HCV (Figure 3).
The proteins of HCV include the structural core and envelope glycoproteins E1 and E2, and the following nonstructural proteins: p7 viroporin and nonstructural protein 2 (NS2) that participate in virus assembly and release; NS3 and NS4A; which is a zinc-binding and proline-rich hydrophilic phosphoprotein and NS3 which is responsible for other NS proteins.73 P7 are viroprions that aid the virus to be released from the host cell after replications through interaction with other viral proteins.77 According to Gailla, C., Tomei, L., & Francesco, R., they have validated that the NS3 protein is responsible for the cleavages of the NS3, NS4A, NS4A-NS4B, NS4B-NS5A, and the NS5A-NS5B junctions (1994).78 While the genomic RNA is translated into a single polyprotein precursor that consists of three structure; the capsid, the Membrane, the Envelope and the seven nonstructural proteins, where only the structural proteins become part of the mature virion, where the polyprotein processes RNA synthesis and virus morphogenesis (Figure 7).79
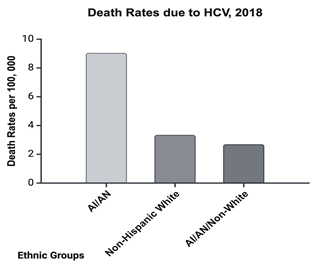
Figure 6 The death rates among American Indians/Alaskan Native per 100,000, compared to non-Hispanic whites, and white Americans. Reported in 2018 by the U.S. Office of Minority Health.
Hepatoviruses genomic variations
There are 5 distinctive types of hepatitis viruses: A, B, C, D, and E, which all infect the liver and cause inflammation, but they share very different genomes. Hepatitis viruses are the leading cause of morbidity and mortality as the consequences of acute chronic infections.80 Hence, HBV and HCV infections account for a substantial proportion of liver diseases worldwide, they have some differences. Like HBV belongs to the Hepadnaviridae family and while HCV belongs to the Flaviviridae family.81 HBV is a double stranded DNA virus, with approximately 3.2 kb and classified into eight genotypes A through H.81 HCV is a single stranded RNA virus whose genome is approximately 9.6 kb, which is quite larger from its counterparts. Hepatitis A Virus (HAV) harbors 7.5 kb genome/polyproteins that is processed into four structural and six nonstructural proteins by proteinase.82 Wassenaar, et al added that, in the lack of the cap assembly that is common in RNA viruses, translation of HAV is initiated by the 5’-untranscribed regions of the RNA genome, which functions as the ribosome entry site (2019).82
HEV, unlike HCV, HEV is a non-enveloped viruses and has a genome length of 7.2 kb, with approximately 33 nm in diameter size, and is a member of the family Hepeviridae83 and contains three virus like proteins (VLP). According to Lemon, M, S., & Walker, M, C.84 in these viruses: HEV and HAV viruses are shed as non-enveloped virions to the environment as compared to their counterparts, while HBV, HCV, and HDV, are the only members that have an envelope. The most fascinating about these hepatitis viruses is that HAV is an RNA virus, as well as HCV, HDV, and HEV, however, Hepatitis B Virus is the only hepadnaviral group that contains a double-stranded DNA virus.85
In this section of the review, we will discuss the different variations in the different Hepatitis viruses. Due to significant genetic heterogeneity, HCV is classified into 7 major genotypes and numerous subtypes that differ in – 30% Nd -20% of their sequences.86 Gottwein et al,86 mentioned that the genotypes also differ biologically and in their rates of response to therapy and antivirals (2011). The 7 genotypes of HCV includes: 1a (isolate H77), 1b (J4 and Con-1), 2a (J6), 2b (J8), 3a (S52 and 452), 4a (ED43), 5a (SA13), 6a (HK6a), and 7a (QC69) which all express recombinants in the core-NS2 proteins (Sheel T et al. 2011). Out of the 7 genotypes, genotype 1a accounts for most of the HCV infections worldwide and were found to be resistant to alpha interferon/ribavirin treatment (Li Y, et al 2014). Of these 7 genotypes, 1a and 2a have worldwide distribution and are known to be associated with different clinical profiling and therapeutic responses (Kato, T et al, 2007).
HCV genotypes and subtypes
Current classification of HCV genotypes in seven major types which include 1, 2, 3, 4, 5, 6, 7,87–89 with genotypes 1 - 3 being widely distributed throughout the world90,91 and genotype 1 being the most common in the United States.92 In few recent studies by Brancho., et al., 2008; Li et al.,18; & Lu L et al.,93 it is explained that of genotype 1 there are seven subtypes (1a, 1b, 1c, 1e, 1g, 1h, and 1i) that were confirmed with genomic sequences. However, Lu L et al.,93. claims that there are 5 additional subtypes of HCV 1 which are 1d, 1f, 1i, 1j, and 1k, that were identified through partial sequencing. Worldwide the most common genotypes are genotypes 1 and 3 and relatively make up about 46% and 30% of HCV cases.94 Keikha et al,94 also states that based on treatment, genotypes 2 and 3 have poor responses as compared to genotypes 1 and 4 (2020).
HCV genotype 3
Within these major genotypes, globally genotype 3 makes up 22 - 30% of all infections and has higher rates of steatosis, faster progression of cirrhosis, and higher rates of hepatocellular carcinoma as compared to HCV genotype 1.95 HCV 3 is the most clinical importance, which confers a high level of resistance to treatments such as daclatasvir and to velpatasvir.96 It is the second most common phenotype in the world, affecting approximately 54.3 million individuals.97 Shahnazariam et al explains that 75% HCV-3 cases occur in East Asia, 54% - 80% in India, 79% in Pakistan, and 30% in Europe (2018).
HCV and BVDV similarities
Surrogate models
Like BVDV, Yellow Fever Virus (YFV) has also been employed as a surrogate model for HCV replication for the evaluation of antiviral agents. Although this virus can also be utilized as another surrogate model, the genome is much more distinct from HCV and BVDV, by harboring a cap-dependent genome instead of an IRES translation. Moreover, BVDV is still the top selection of HCV surrogate model because of its noninfectious status against humans, while YFV has mostly been used for evaluating compounds for antiviral efficacy. There have been reports on a surrogate model that were used to develop an antiviral drug for Hepatitis E viruses. Debing Y et al.,101 and his colleagues employed cutthroat trout virus (CTV), which is a nonpathogenic fish virus with remarkable similarities to HEV, and as a potential surrogate for HEV and established an antiviral assay against this virus using the chinook salmon embryo (CHSE-214) cell lines.
Discovery of microRNAs
Since the discovery of the first microRNA (miRNA) in Caenorphabditits elegans (C. elegans), it has changed the field of molecular biology. MicroRNAs (miRNA) are a class of small non-coding RNAs.102–105 These molecules play crucial roles in cell replication, differentiation, immune responses, viral replication and have been used for antiviral drugs and vaccine development. Many of the miRNA known today, have been uploaded and listed in computer user friendly databases that can be easily accessible to users (Figure 8) (Figure 9).
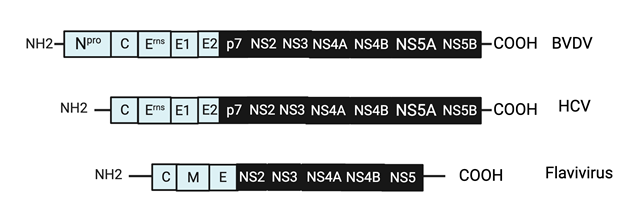
Figure 8 Comparison of the viral polyprotein genomes of BVDV to HCV, and BVDV, HCV, to the genomic polyprotein of the flavivirus family.

Figure 9 Nucleotide sequence alignment, generated by Mega alignment database. The BVDV type 1 nucleocapsid sequences aligned with the Capsid/Core sequences of the HCV virus and its 6 genotypes. Perfect alignments are indicated by *.
MicroRNA roles and functions
Unlike mRNAs, which encode proteins, miRNAs do not but they do control various levels of gene expressions.106 There are three type of small non-coding RNAs: small interference RNA (siRNA), micro RNAs and RNA interference (RNAi), that are considered as a significant posttranscriptional gene silencing mechanism and functions as an antiviral immunity in eukaryotes.107 Zhao, Y., & Srivastava, D., also classified additional miRNAs: the repeat-associated small interfering RNAs, and piqi-interacting RNAs (piRNAs) (2007).108 They are critical for development, used as biomarkers for diseases, and regulate cell-cell communications.109 miRNA are small RNA molecules with length of 18 to 25 nucleotides (nt) and have been detected in many plants and animal species and even in some animal viral RNA genomes.110 miRNAs sequences expressed from longer transcripts encoded in animal, plant, and virus genomes and recently discovered in single-celled eukaryotes.111 Griffiths-Jones et al continues that miRNAs also regulate the expression of target genes by binding to complementary sites in transcripts and cause translational repression or transcript degradations (2007).111
The Review also reveals various databases that are considered useful in developing and annotating miRNA sequences for specific targets. Examples of databases that contain miRNAs are miRDbase, microRNA REGISTRY, NCBI Basic Local Alignment Search Tool (BLAS), MiRBase Targets, and miRanda Algorithms. With the use of these databases miRNAs can be sequenced, aligned, and amplified using alignment search tools. One of which was presented by Griffiths-Jones, et al.104 who used the miRBase database which is aimed to provide integrated interfaces to comprehensive miRNA sequence data, annotation, and predicted gene targets. miRBase targets is a comprehensive new database of predicted miRNA target genes at http://microrna.sanger.ac.uk.104 This review will also discuss the impact of HCV on the indigenous populations, genomic similarities within HCV and BVDV, and miRNA’s role and functions in vaccine development against these viruses.
miRNAs are known to have a major role in regulating the expression of protein-coding genes, to block invading viruses and to quell the genomic incorporation of genetic parasites like retroviruses and lentiviruses.112 miRNA may be capable of inhibiting viral replication, they are also known to enhance the virulence of viral replication. For example, miR-122 is a liver-specific miRNA and most abundant in the liver (70%) that positively regulates HCV RNA abundance and essential for production of infectious HCV (Jangra et al., 2010). miRNA are also crucial regulators of innate and adaptive immune responses by activating T and B cells and their abnormal expression function of the immune system have been linked to multiple human’s diseases including inflammatory disorders such as inflammatory bowel disease, and cancers.113
MicroRNA inhibitors against various viruses
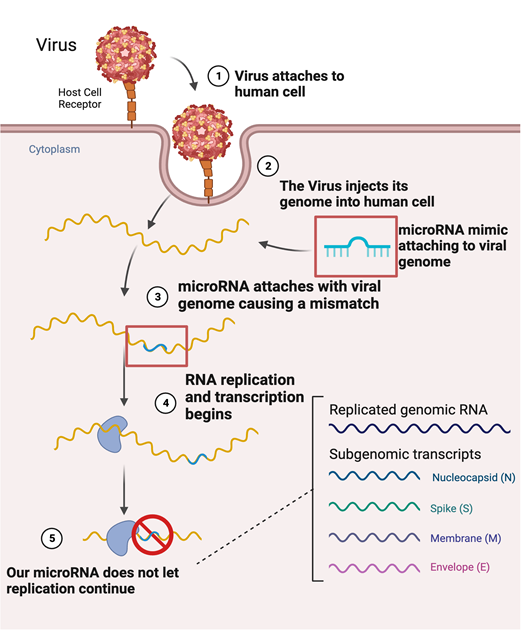
Figure 10 Mechanism of inhibitions of microRNAs attaching to specific parts of a viral genome causing a mismatch, leading to the inhibition of the viral genome.
There was also data available that validated that miRNAs were used against flaviviruses and used as inhibitors against their replication cycle. In an article written by Saha, A et al, they used an artificial miRNA (amiRNA) based approach by using vector-delivering amiRNA to effectively inhibit viral replication of Chikungunya virus (CHiKV) by inhibiting the E2 protein (2015). The literature also demonstrated that several RNAi were used to reduce the viral protein expression in the core and E2 protein of HCV, because the core and the E2 protein play crucial roles in viral infection and replication cycle.122 In the experiment it mentioned the use of four miRNAs that were used against the two E2 proteins and two C proteins and show inhibitory effects of HCV. This discovery has led the development of antiviral therapeutics against HCV in humans.
According to Slonchak et al,123 they have used a miRNA called miR-532-5p, which was used against the viral replication in WNV. The mentioned miRNA has enhanced virus infection by 8 – folds at 24 hpi but inhibited viral replication approximately 10-fold at 48 hpi. As of late, miRNAs have been used against majorities of viruses related to the flaviviruses, but they were also effective against other viruses as well such as the Flu, Rabies, CHiKV, FMDV, and members of the Flavivirus families, data available in various literature has also demonstrated the use against miRNAs that even inhibited the replication of COVID-19.
Overall, this review article discussed the impacts Hepatitis C has in both the United States, and globally, but also on the indigenous communities, and how it still remains a threat to public health. With no suitable cell culture system and no production of a vaccine, HCV remains a major risk factor for the development of liver cirrhosis and hepatocellular carcinoma.124 Buckwould et al.124 explains that without an authentic method to grow and maintain Hepatitis C in cells, surrogate viruses are continuously being utilized. Without a reliable cell culture system, the viral life cycle remains complex and not fully understood.125 While there are some differences, BVDV shares similarities in replication cycle, biology, and genetic organization with Hepatitis C, and hence BVDV is mainly used as a surrogate model for in vitro testing in the search of antivirals against Hepatitis C.126
Even with the discoveries of using the JFH1 method to study Hepatitis C in vitro, there is still no cell culture that can maintain all genotypes, meaning that development of a novel cell culture system is still a needed priority. Although people have cloned full length Hepatitis C genomes and confirmed their infectivity in chimpanzee models, none of them is infectious in cell cultures except for the JFj-1, which is the first clone to support efficient cirrus production in Huh7 hepatoma cells.93 Even though that JHF1 was proven effective, the sensitivity of the different genotype viruses were relatively small127 and the titers of viral RNA level are relatively low.128
Though miRNAs have been discovered in invertebrates, there are miRNAs found in plants too, which could also play a possible role in antiviral mechanisms.129 This review also discusses the importance of miRNAs, how they play pivotal roles in biological processes including cell proliferation, metastasis, differentiation, development and apoptosis.130 They were even shown to have the capabilities to block viral replication for virus infections. These miRNAs have additional roles other than the already mentioned functions, they are important for natural target recognition.131 It has mentioned that the host miRNAs were shown to interact directly with viral RNAs of RNA viruses and modulate replication (Sonchak et al 2016). They can also silence genes and block replication in various viruses, which can be used as an antiviral therapeutic and used as vaccines. In validation that RNA viruses do have miRNA-binding sites within their genome and miRNAs can bind to RNA virus genomes, enhancing genome stability, repressing translation and altering free miRNA level in the cell.132–145
The Flavivirus family consists of positive strand RNA viruses and includes viral genus pestiviruses and hepatoviruses. HCV is a member of the same family that remains a threat to public health due to the unavailability of appropriate vaccines to prevent HCV infection. BVDV, a member of the pestiviruses genus and a member of the flavivirus family, has proven to be related to Hepatitis C, which is why they are used as surrogate models. With the use of miRNAs, which are small noncoding RNA molecules ranging from 18 – 25 nucleotides that form a hairpin structure, they play an important role in gene expression, cell proliferation, cell apoptosis, enhancing viral replications, and are used as target genes against viral mRNAs for development of vaccines and antiviral synthesis. With the information presented in this review paper, we anticipate possible use of these miRNAs that could target and inhibit the BVDV replication process of the nucleocapsid protein, and later could potentially use to inhibit HCV.
The authors gratefully acknowledge the moral support from School of STEM, Diné College and financial support from MS Biology Grant # NSF-TCUP-ICE-TI-2202023 (PI: Dr. Donald K. Robinson).
Author contribution
Harrison Cayatineto is the main author of this article which is part of his MS project. Dr. Hakim has contributed by providing guidance in writing this review, timely feedback and editorial work.
National Science Foundation (NSF) - NSF-TCUP-ICE-TI-Grant #2202023.
The authors declare that there are no conflicts of interest.

©2024 Cayatineto, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.