Journal of
eISSN: 2373-633X


Research Article Volume 9 Issue 4
1Leader researcher, Cancer Research Center, Russia
2Department of Physics, Moscow State University, Russia
3Researcher, Cancer Research Center, Russia
4Head of Laboratory, Russia
Correspondence: Anna A Lushnikova, N N Blokhin Cancer Research Center, 115478Kashirskoye shosse-24, Moscow, Russia
Received: July 25, 2018 | Published: July 27, 2018
Citation: Lushnikova AA, Kostarev AV, Onyan AV, et al. Simulation binding between nucleoli and cationic peptides, inducing tumor cell apoptosis, by molecular docking. J Cancer Prev Curr Res. 2018;9(4):187-189. DOI: 10.15406/jcpcr.2018.09.00347
Backgound: cationic peptides and their possible targets─chaperone proteins nucleolin, NCL and nucleophosmin, NPM, regulating key cellular processes, are promising as antitumor agents. Differential expression of NCL and NPM, especially cell surface nucleolin, in tumor and normal cells creates the basis for selective cytotoxicity of the peptides and their clinical application.
Aim: objective is to propose strategy for analysis binding between nucleolin and peptides by molecular docking.
Results: nucleolin structure is rather stable and cross-linking between NCL and peptides affects amino-acids with constant positions in NCL moleculae. Cationic peptides are suitable as ligands for nucleolin dimer.
Keywords: tumor cells, nucleolin expression, cationic peptides, target-ligand docking
Chaperones nucleolin, NCL and nucleophosmin, NPM are multifunctional proteins involved in the regulations of gene transcription, RNA synthesis, translation and metabolism, ribosome biogenesis, chromatin remodeling, control of cell proliferation and apoptosis, molecular transport and other important processes.1 These proteins over-expressed in highly proliferative tumor cells and localized presumably in the nucleolus. It has also been revealed that glycolsilated and phosphorylated nucleolin is highly expressed on tumor cell surface as receptor for different ligands to trigger molecule internalization, including potential anti-cancer agents–cationic peptides.2 The mechanism of binding between receptor nucleolin and peptide molecules is unclear; its analysis by molecular docking has been considered.
Molecular structure, representing NCL RRM_1 domain─RNA-recognition motif, taken from PDB (ProteinDataBank 2FC8), was used as a target protein. It was obtained by NMR (SOLUTION NMR – Solution nuclear magnetic resonance spectroscopy, Nuclear Magnetic Resonance).3 It includes 1 protein chain, 743 atoms, 102 amino-acid residues. We used the program Maestro 11. First stage of the protein preparation consisted of protonation, pH=7.0 by water removing and single minimization of structure using force fild OPLS3, because a larger number of minimizations might lead to the protein chain deformation.4
Two problems had to be solved before molecular docking:
The docking was performed by glide.6 In total, 4 configurations for each conformer and score function GS per the configuration were calculated. The results of the screening for these structures are shown on Figure 1. Interestingly, that majority of the peptide fragments had a high score >6.00kcal/mol (value modules). All these structures located in approximate active center of the protein, and formed hydrogen bonds between the same amino acids. This fact suggests the idea on the characteristic regions of binding between peptides and target protein. To verify this estimation, the best structures with position scores more than 6.50 kcal/mol (value modules) were re-positioned, provided that the binding areas remain mobile. In this case, the scoring function changes and the protein conformation changes is partially added to it. Then site will be selected for the ligand followed by its positioning into this site. Such approach may give the best result for estimation of the binding between protein and ligand.7 Before induced fit docking procedure described in,8 we have identified amino acids that are commonly contacted with the leaders of the first docking. The frequencies of the relationships between the ligand with leader score and appropriate amino acid were compared. Five amino acids in nucleolin molecule binding peptides more frequently, were revealed (Table 1).
Amino acid in NCL molecule |
Binding frequency with ligand,% |
GLU 29* |
71 |
ARG 43 |
71 |
THR 46 |
43 |
ASP 60 |
71 |
ASP 87 |
100 |
ALA 89 |
29 |
LYS 90 |
71 |
PRO 91 |
29 |
Table 1 The frequencies of hydrogen bounds between ligands and NCL,*- amino acid position
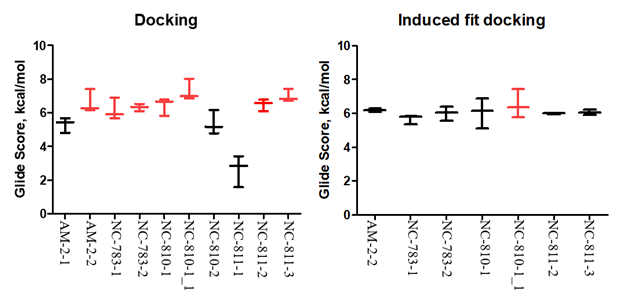
Figure 1 Resulting diagrams on molecular docking and Induced fit-docking.
Based on the selection results, the program changes a spatial arrangement of the amino acid that forms hydrogen bounds with a frequency >71%. Docking box was also centered according these amino acids and the box size of 26 Ǻ followed by its reducing, since the binding site or active center has been already detected.In summaty, the score functions did not change, they were preserved close to the previous values. Visually assessing the amino acid changes, one can agree, that the ligand binding site within the target protein does not undergo significant changes. This observation indicates that nucleolin’s active center itself has a rigid structure and binding is not related to the mobile, changed sites, but remains relatively stable.
Nucleolin is a multifunctional chaperone protein over-expressed in tumors, especially on cell surface. It considered now as a strategic target for non-invasive tumor imaging, cancer diagnosis and for targeted delivery of the drugs and triggering inhibition of tumor growth by different agents, for instance, cationic peptides. Simulation binding between four cationic peptides and nucleolin using molecular docking approach, has revealed, that these peptides are suitable as nucleolin ligands (Figure 2). Moreover, preferable binding sites with stable position and high glide scores were observed in nucleolin dimer 3D-structure. The data would be a base for study of cationic peptide toxicity to tumor cells and design optimal peptide structures with anti-cancer activity.
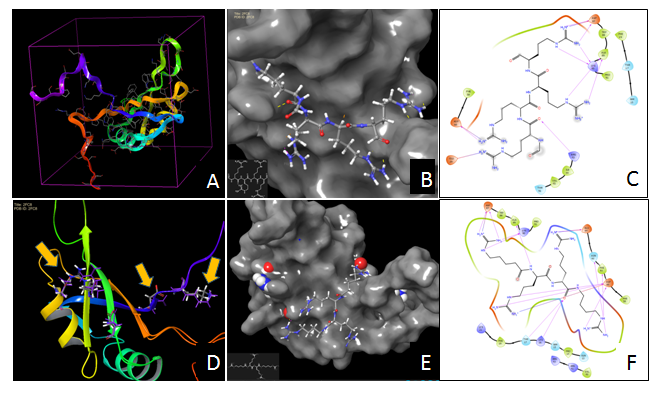
Figure 2 Molecular docking of peptide fragments into nucleoli active site.
None
No conflict of interest between the authors.

©2018 Lushnikova, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.