Journal of
eISSN: 2373-633X


Review Article Volume 3 Issue 3
Department of Biomedical Science, Estÿcio do Amazonas University, Brazil
Correspondence: Thais Sobanski, Biomedical Science Student, Estÿcio do Amazonas University, Av Constantino Nery, n?? 3693, Bairro: Chapada CEP 69.050-002, Manaus, Brazil, Tel 55 92 994169469
Received: July 25, 2015 | Published: October 30, 2015
Citation: Sobanski T, Fraporti L, de Almeida TAP. Proteins involved in DNA double-strand breaks repair pathways are essential to prevent the development of cancer. J Cancer Prev Curr Res. 2015;3(3):278-282. DOI: 10.15406/jcpcr.2015.03.00081
The single-stranded DNA binding (SSB) proteins play essential roles in the repair of many types of DNA damage, including double-stranded breaks (DSBs). Double-strand breaks (DSBs) are one of the severest types of DNA damage. The single-stranded binding protein is also important to maintain genome stability, since unrepaired DSBs easily induce cell death or chromosome aberrations. To maintain genome instability, cells have developed a cell-intrinsic network mechanism called DNA Damage Response (DDR) throughout most of the cell cycle. There are two main pathways of DSBs repair mechanisms, non-homologous end joining (NHEJ) and homology directed repair (HR). In this perspective, we will describe how single-stranded DNA binding proteins functions during the DSB repair pathway and their consequences for genome stability and cancer.
Keywords: single-stranded DNA, double-strand breaks, DNA damage response, pathways, cancer,, genetic material, deamination, homologous recombination, ultraviolet, ionizing radiation, hypomorphic mutations, humoral immune deficiencies, immune response, radio sensitivity, adenylation
SSB, single-stranded DNA binding; Dsbs, double-stranded breaks; DDR, DNA damage response; HR, homologous recombination; NHEJ, non-homologous end joining; UV, ultraviolet; IR, ionizing radiation
In the last few decades, cancer research has gained remarkable insights showing that cancer is a genetic disease. Damage to genetic material is a persistent and ubiquitous threat to genomic stability. To transmit genetic information from one generation to the next, it is essential that DNA is protected from the damage caused by environmental agents and by that produced during DNA metabolism. Cells continuously encounter DNA damage from either endogenous sources including radical species as by-products of cellular metabolism or from exogenous sources. Endogenous DNA damage can lead to DNA alteration during replication; inter conversion between DNA bases generated by deamination or loss of bases following DNA modification in a process called alkylation. In addition, oxidized DNA and DNA breaks can be generated by oxygen free radicals resulting from normal cellular metabolism. Exogenous agents can be classified as air pollution, cigarette smoke, food additives, toxins, and ultra-violet rays in sunlight.1–4 For example, physical genotoxic agents such as ionizing radiation (IR) and ultraviolet (UV) light are estimated to induce 105 DNA lesions, such as chromosomal breakage (pyrimidine dimer and 6-4 photoproducts) per cell a day (Figure 1).4
There are two types of DNA strand breaks that can occur; when the lesion is just in one of the two strands, single strand breaks occur. However, when two of these breaks are close and on opposite strands it is classified as a DNA double-strand break (DSB).5 DNA DSB double-strands breaks (DSBs) are an extreme threat for genome integrity because they can lead to chromosomal rearrangements or loss of genetic material. Almost all human cancers arise as a result of genomic instability that drives the carcinogenesis process.
To monitor the genome integrity, cells have developed a cell-intrinsic network mechanism called the DNA Damage response (DDR) and failure of this process often results in apoptosis or genomic instability, such as aneuploidy, deletion, or trans- location.
DNA damage response (DDR) is divided in three main steps
The DDR recognizes DNA lesions and initiates various downstream pathways, including cell-cycle arrest, transcriptional and post-transcriptional activation of a subset of genes associated with DNA repair, and under some circumstances triggers programmed cell death.6 There are four distinct mechanisms of DNA DSBs repair in mammalian cells that have been classified as: non-homologous joining (NHEJ), alternative-NHEJ, single strand annealing and homologous recombination (HR). NHEJ and HR are the two major DNA DSB repair pathways.7
Double strand-breaks: the two main mechanism of DNA repair
In mammal’s cell, there are two main mechanisms of double stranded-DNA breaks repair: homologous recombination (HR) and non-homologous end joining (NHEJ).8 Homologous recombination is an error-free repair mechanism that utilizes the genetic information contained in an undamaged sister chromatid as a repair template. Also, this mechanism predominantly operates in the late S and G2 phase of the cell cycle, since it is when the chromatid sister is available as a template and this pathway is considered a more precise to repair DSBs in DNA;9 Otherwise, NHEJ is often error-prone pathway and involves elimination of DSBs by direct ligation of the broken ends and this mechanism canoperates throughout the cell cycle. In addition, this mechanism tolerates nucleotide (nt) loss or addiction during the rejoiningsite (Figure 2).10
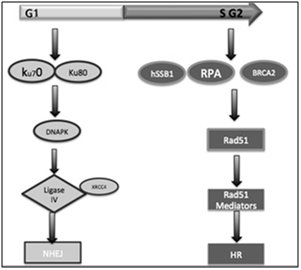
Figure 2 Homologous recombination is an error-free repair mechanism that operates in the late S and G2 phase of the cell cycle, since it is when the chromatid sister is available. While, NHEJ can operate mainly during G1 phase or throughout the cell cycle.
Non-homologous end joining (NHEJ) pathway
NHEJ can operate throughout the cell cycle, but it has been shown to predominate mainly during the G1 phase.11 In mammalian cells, NHEJ initiates with a limited processing of DNA ends by the MRN complex composed of the meiotic recombination (MRE1), (RAD50) and (NBS1); also known as nibrin proteins, being considered as a central protein complex to recognise the DNA breaks.12 When the DSBs are recognized and processed by MRN complex a signaling cascade begins which allow proteins ku-70and ku-80 binds at the DNA ends and recruit the DNA dependent-protein kinase catalytic subunit (DNA-PK). This interaction between Ku70/80 and DNA-Pk play an important rule to synapse the two DNA ends to be repaired. Also, Ku70/80 interaction seems to improve the binding equilibrium of enzymes, such as nucleases, polymerases, and ligase. These enzymes employee a sophisticated engineered machine to align a pair of ends together and perform the ligation step. Following that, once DNA-PKcs bounds to brokes ends, it seems to activate the serine/threonine kinase of this complex representing a simple’s signal transduction since it allows DNA-Pkcs to phosphorylate itself causing conformational changes. Artimis is also an important enzyme to function as a 5’-3’ endonuclease and these conformational changes seems to help recruiting two of known polymerases μ and λ for the NHEJ complex.13 A complex formed by XLP, XRCC, and DNA ligase IV composes the ligation of DNA ends. The function of XRCC is stabilizing ligase IV protein in cell improving its enzyme activity and efficiency of the adenylation.
The protein XLF stimulates XRCC4binds to DNA ligase IV in the presence of divalent cation Mg.14,15 To understand this DSBs breaks pathway repair is essential since it contains many proteins that could be target to improve patients’ outcomes or eliminate cancer. During the treatment, therapeutic agent’s causes DSBs breaks in the genome as an intermediate and inhibitors could be used to block this residual process. Therefore, targeting therapy against the key signaling molecules has therapeutic implications (Figure 3).
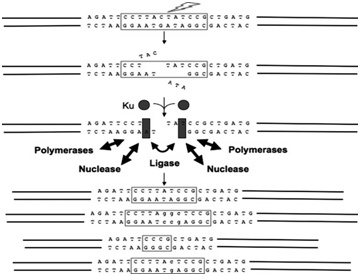
Figure 3 DSBs are recognized and processed by MRN complex and a signaling cascade begins allowing proteins ku-70and ku-80 binds at the DNA ends and recruit the DNA dependent-protein kinase catalytic subunit (DNA-PK).
Human single-strand DNA binding protein - the key to genomic integrity
Single-stranded DNA (ssDNA) binding proteins play acentral rules in DNA during replication, recombination, DNA damage signaling, and repair in all living system. Single-stranded-binding proteins (ssDNA) have an efficient mechanism that operates in a sequence-independent manner protecting the DNA from chemical and nucleolytic attacks.16 hSSB1 (human single-strand DNA binding protein1) is a recently discovered protein that has high affinity to the single- strand DNA molecule in human cells.17 hSSB1 protein has been shown to be critical for the maintenance of genomic stability since it is involved in the precise repair of double-strand breaks (DSBs) by homologous recombination (HR). The human single-stranded (ssDNA binding complex, such as hSSB1) is involved in the regulating DSB signaling and HR repair. hSSB1 is an essential protein to initiate homologous recombination since it has the ability to bind single strands DNA generating a complex signaling pathway. The occurrence of double-strand breaks (DSB) by exogenous or endogenous agents generates single strand DNA (ssDNA) and hSSB1 recognizes these breaks. The hSSB1-ssDNA binding is important to recruits MRN complex to the DNA breakage site. The Mre11, Rad50 and two NBS1 polypeptides, where the C-terminal hSSB1 protein only interacts directly with the N-terminus of the NBS1 protein.18 Homologous recombination initiates when it is processed by MRN complex through 5’ strand at DSB ends to produce a 3’single-stranded DNA (ssDNA). The resulting 3’ ssDNA tails is rapidly bound by replication protein A (RPA); this association has the function to protect the DNA from it damage and digestion and preventing formation of disruptive secondary structure.19 Following that, the RPA-ssDNA complex is displaced by Rad51 protein, a key recombinase enzyme, and the displacement of RPA complex is important for completion of homologous recombination events. The loading of Rad51 is mediated by BRCA2 generating a nucleoprotein filament allowing strand invasion. This event permits the DNA strand exchange occurrence generating a joint molecule between damaged and undamaged duplex DNAs. The DNA information that is lost at the damaged DSB breakage site is recovered by DNA synthesis using an un damaged homologous strand as a template. Finally, the stand invasion formation is intermediated and processed by branch migration, Holliday junction and DNA ligation.20 Among these several steps that are present during Homologous Recombination, the recognition of DSB by hSSB1 and the recruitment of MRN complex are essential for cells successfully repair DNA Double- strand breaks (DSBs) (Figure 4).
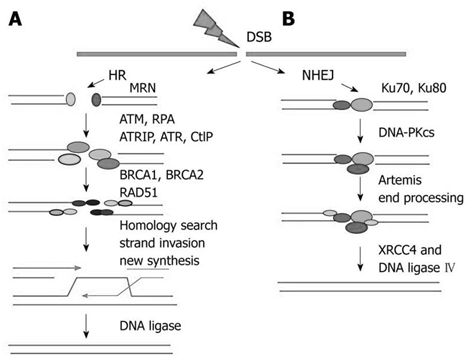
Figure 4 DNA double-strand breaks (DSBs) can be repaired by homologous recombination (HR), which requires a homologous sequence as a repair template, or by end joining, which involves either more-or-less direct ligation of D.
Dsbs proteins defects associated with human syndromes and increased cancer risk
DNA double strand breaks are constantly generated in our cells either through external agents or through internal agents, including sub products from cellular metabolism. In order to maintain the genome stability cells have developed an efficient surveillance network that can detect and repair DNA breaks to protect the development of cancer or human syndromes caused by mutation of genes coding for DSB signaling and repair pathways proteins. These syndromes associated with mutations of these binding proteins involved in DSBs repair share many clinical similarities, including neurological defects, growth delay, immunodeficiency, radio sensitivity, sterility and increased cancer incidence. Increasing the knowledge on these proteins involved in DSBs repair can provided great insight into the physiological functions of DSB response proteins, which may led to rapid discoveries of knew proteins that can be targeted to drugs development. Therefore, the single-stranded DNA binding (SSB) proteins is critical to prevent pathologic chromosome rearrangements and subsequent tumor development.
The authors thank the Translational Research Institute (TRI), localized in Brisbane, AUS for offering a better understanding how normal human cells loose genomic stability and how these DNA repair pathways become aberrant in cancer. Also, would like to thank Cnpq, which offered the opportunity to make part of the program called Science without Borders. In particular, I would like to thank Dr. Felipe Gomes Naveca and MSc. Tatiana Almeida Pires for the countless ways they have supported, mentored, and motivated me during my Biomedical Science Bachelor.
Contributions
Liziara Fraporti and Tatiana Amaral Pires de Almeida contributed to: study design, analysis and interpretation, manuscript preparation, manuscript re-editing and manuscript review.
The authors declare there is no conflict of interests.
None.

©2015 Sobanski, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.