Journal of
eISSN: 2373-633X


Research Article Volume 8 Issue 5
1College of Medicine, University of Al-Qadisiyah, Iraq
2Department of Chemistry, College of Education, University of Al-Qadisiyah, Iraq
Correspondence: Huda AlSayed Ahmed, PhD, Clinical Laboratory Specialist, Johns Hopkins Aramco Heathcare, Dhahran, KSA, Tel 9665803854
Received: August 14, 2017 | Published: October 24, 2017
Citation: Jabir FA, Hoidy WH. Pharmacogenetics as personalized medicine: association investigation of SOD2 Rs4880, CYP2c19 Rs4244285 and FCGR2A Rs1801274 polymorphisms with breast cancer population in Iraqi women. J Cancer Prev Curr Res. 2017;8(5):348-353. DOI: 10.15406/jcpcr.2017.08.00294
Breast cancer is the most common cancer in women characterized by a high variable clinical outcome among individuals treated with targeted therapies. In this study, we performed a population based approach intersecting high-throughput genotype data from Iraqi populations with publically available pharmacogenomics information to estimate the frequency of genotypes correlated with responsiveness to breast cancer treatment thus improving the clinical management of this disease in an efficient and cost effective way. A total of 50 patients and 25 healthy controls were enrolled in our study. Genotyping of rs4880, rs4244285and rs1801274, was examined association with breast cancer in Iraqi women. We found that individuals carrying the CT genotype of rs4880manifested an increased risk of breast cancer in comparison with those carrying the TT genotype (OR=0.171, 95%Cl=0.053-0.551, P=0.002). in dominant model, we observed that CT&CC genotype of rs4880 showed an increased risk of breast cancer compared with the TT genotype (OR=0.248, 95%Cl=0.089-0.690, P=0.006). Moreover, subjects with the GA genotype of rs424485 presented a high risk of breast cancer than the GG genotype (OR=0.256, 95%Cl=0.066-0.987, P=0.038) and dominant models (OR= 0.025,95%Cl=0.054-0.775, P= 0.013). The analysis revealed that rs1801274 showed linkage disequilibrium and decreased risk with breast cancer. In conclusion, our study suggests that rs4880and rs4244285 polymorphisms play an important role in development of breast cancer in Iraqi population, and no significant association between rs1801274 and breast cancer.
Keywords: breast cancer, pharmacogenetics, personalized medicine, SOD2, CYP2C19, FCGR2A
Breast cancer is the most frequent carcinoma in females and the second most common cause of cancer related mortality in women.1 It is the most commonly diagnosed malignancy in women around the world, especially in the Western countries. It accounts for almost one fifth of deaths caused by cancer.2 Every year, one million new cases are reported worldwide, representing 18% of the total number of cancer in women. It has been established that one out of eight women (in USA) and one out of 10 women (in UK),3 will develop breast cancer at some point in her life.4 The death rate for breast cancer has been slowly declining over the past decade, and the incidence has remained level since1988 after increasing steadily for nearly 50 years. Twenty-five percent to 30% of women with invasive breast cancer will die of their disease.5 But this statistic, as grim as it is, also means that 70% to 75% of women with invasive breast cancer will die of some thing other than their breast cancer. Hence, a diagnosis of breast cancer, even invasive breast cancer, is not necessarily the ‘‘sentence of death’’ that many women imagine.
Mortality rates are highest in the very young (less than age 35) and in the very old (greater than age75). It appears that the very young have more aggressive disease, and that the very old may not be treated aggressively or may have comorbid disease that increases breast cancer fatality.6 In Iraqi, where the population was exposed to high levels of depleted uranium following the first and second gulf wars. Breast cancer is the most common type of malignancy among the Iraqi population in general; responsible for about one third of the registered female cancers and almost one quarter of female deaths from the disease. Within the last two decades, there has been an obvious increase in the incidence rates of breast cancer, which became one of the major threats to Iraqi female health.7
Pharmacogenetics has been defined as the science of pharmacological response and its modification by hereditary influence; the subjects of interest relate both to efficacy (therapeutic effectiveness) and toxicity (production of side effects or unwanted effects).8 Dealing with the genetic basis which underlies variable drug response in individual patients, it develops an individualized approach to the therapy, where optimally effective drugs are matched to a patient's unique genetic profile.9 Pharmacogenetics provides insight into the molecular level of drug function and, as a consequence, offers the potential of individualized drug therapy.10 Thus it can help in optimizing drug efficacy and minimizing adverse drug reactions.11
The term pharmacogenetic polymorphism defines a monogenetic trait, caused by the presence of more than one allele at the same locus and more than one phenotype in the same population in regard to drug interaction with the organism, with the frequency of the rarest allele of more than 1%. DNA sequence variations may occur as insertions or deletions of nucleotides, differences in the copy number of repeated sequences or SNPs - Single Nucleotid Polymorphisms.12 If the mutations occur in proteins that are drug targets or drug-metabolizing enzymes, or in proteins that are involved in drug transport mechanisms, they can affect drug efficacy and safety.13
Anthracycline-based adjuvant regimens have become the standard of care for early-stage breast cancer in the United Kingdom. The regimen of doxorubicin and cyclophosphamide (AC) is one of a number of available choices, with widespread use in patients with an indication for chemotherapy, but a low to moderate risk of recurrence. The combination of doxorubicin and cyclophosphamide was first tested by the National Surgical Adjuvant Breast and Bowel Project as a simple alternative regimen to replace cyclophosphamide, methotrexateand5 fluorouracil (CMF), which had been established as an effective adjuvant treatment.14
Cyclophosphamide is a prodrug activated by a number of different cytochrome P450 enzymes, including CYP2C19.15 Each of these enzymes and transportergenes is known to exhibit a degree of genetic variation, characterised by single nucleotide polymorphisms (SNPs). These SNPs are present at significant frequencies in a European population and their influence on the pharmacology of a number of different agents has been characterised.16
The aim of this study was to investigation of SOD2 rs4880, CYP2C19 rs4244285 and FCGR2A rs1801274 polymorphisms with breast cancer population in Iraq women and examine the possible influence of SNPs on the tolerance, side effects and overall clinical outcome of chemotherapy in patients with breast cancer.
Patients and clinical samples: the blood samples from 50 patients with different stages of breast cancer were provided by Tumor center/ Al-dewanyah teaching Hospital /AL-Dewanyah during the period from May 2016 to December 2016. 25 blood samples from healthy donors were used as a control in this study. From each sample, 2 mL of peripheral blood was collected into an EDTA-containing tube. The samples were stored at -20Cº until further processing. The main data and parameter include in our study: patient’s age, family history of breast cancer and another cancer, and blood group
Measurement of DNA Concentration and Purity
Extracted DNA concentration and purity were measured by the nanodrop (Quawell/Hong Kong), 2µl of DNA was loaded to the lens of the nanodrop and measured on 260/280 nm wave length, the result appeared on the laptop screen that attached to the nanodrop.The nanodrop lens was cleaned by distilled water (D.W) and cotton swap after each sample, the other samples were measured consequently.
Primers
Primers that used to identify and amplify the CYP2C19, rs4244285. SOD2, rs4880 and FCGR2A, rs1801274 fragment is given in Table 1.
|
Product size |
Tm (ºC) |
Sequence (5 - ˃3’) |
SNP Name |
|
408 |
54 |
F 5- TTACAACCAGAGCTTGGCAT -3 |
rs4244285 |
|
56 |
R 5- TCCTTGACCTGTTAAACATCCGT -3 |
||
|
400 |
60 |
F 5- GAGGGGACGCGGGGA-3 |
rs4880 |
|
60 |
R 5- CCCTGGGGTCGCCTCT-3 |
||
|
420 |
57 |
F 5- CTTGGCAGACTCCCCATACC -3 |
rs1801274 |
|
|
55 |
R 5- AGTTCTGTGAGTAACGTACCTCTG -3 |
|
Table 1 Sequence of primers used in the present study
Polymerase chain reaction (PCR)
PCR technique was used to identify and amplify the 408 bp band of CYP2C19 gene, by using the primers that designed by Alfa Gene/Canada (Table 2). The primers were lyophilized, they dissolved in a free ddH2O to give a final concentration of 100 pmol/µl (as a stock solution) and kept as a stock in -20Cͦ to prepare 10 pmol/μl concentration as work primer resuspended 10 μl of the stock solution in 90 μl of free ddH2O to reach a final volume 100μl (10 Pmol).
|
Component |
Reaction size |
|
Template DNA |
5 µl |
|
Primers(10 pmol/ml) |
1µLF |
|
1µLR |
|
|
Deionized Water |
13µl |
Table 2 PCR reaction components for amplification of the 408 bp band
The total volume of PCR reaction was 20µl, the reaction components are described in Table 2. Then PCR product was separated on 2% agarose gel at 50 volt for 45 min in 0.5X TBE buffer by taking 5 µl from each sample, agarose gels were stained with ethidium bromide 0.5mg/ml for 20-30 minutes. DNA band was visualized by electrophoresis and captured by gel documentation system to document the observed bands.
Statistical analysis
Genotype and allele carrier frequencies were defined as the percentage of individuals carrying the genotype and allele out of the total number of individuals, The χ2 and (p value<0.05), the odds ratio (OR) and their confidence intervals (95% CI) tests of SPSS 23.0 for Windows (SPSS Inc., Chicago, IL, USA) were used to compare the frequency of discrete variables between patients with breast cancer and control individuals.
The risk of breast cancer with some variables such as smoking and lodging were estimated by the odd ratio and their confidence interval at (p value less than 0.05), the matching in ages between cases and controls was demonstrated by p value by using the SPSS software as shown in Table 3. In our study and as obvious from the results above there is no association between lodging and smoking with risk of breast cancer (p value was higher than 0.05).17
|
Variables |
Control (G1) |
Patients (G2) |
OR (95% CI) |
P Value |
|
N= 25 (%) |
N= 50 (%) |
|||
|
Logging |
||||
|
Urban |
18 |
32 |
0,691(0.243-1.969) |
0.488 |
|
Rural |
7 |
18 |
||
|
Smoking |
||||
|
No Smoker |
23 |
42 |
2.190(0.492-11.188) |
0.337 |
|
Smoker |
2 |
8 |
||
|
Age (mean ±SE) |
29.42±10.21 |
51.29±12.18 |
|
|
Table 3 Clinical characteristic and breast cancer risk
Genomic DNA of sufficient quality and quantity was extracted from 50 blood samples from women with breast cancer and 25 women as healthy ( control ) from AL_Diwanyah teaching hospital, Amplification of the SOD2 rs4880, CYP2C19 rs4244285 and FCGR2A rs1801274 target sequence from these archival samples resulted in 400, 408 and 420 bp products, respectively. The amplified fragment which is yielded of single band of the desired product with a molecular weight of genes appeared sharp in agarose gel through gel electrophoreses technique and loaded with (25-1000bp) DNA ladder (Figure 1).
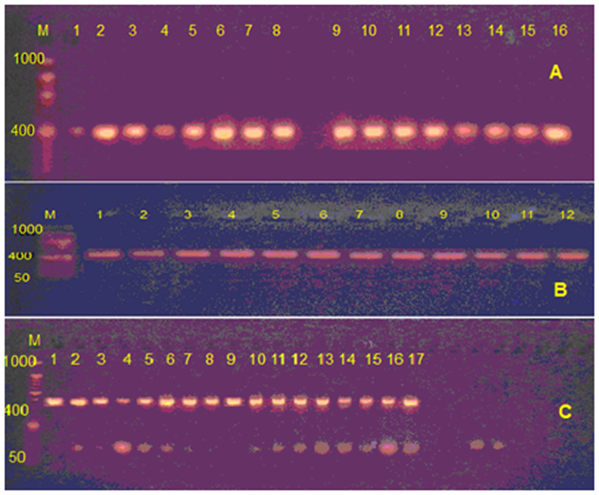
Figure 1 Pcr product was electrophoresis on 2.5% agarose gel at 50 volt, 45 min: M refers to (25-1000 bp ladder). A refer to rs4880 for SOD2, B refer to rs4244285 for CYT2C19 and C refer to rs1801274 for FCGR2A.
Seventy five PCR product samples were sent for sequence analysis; 20µl of PCR product for each sample was sent and 100 µl (10 pmol) from the forward primer. DNA samples were sequenced using AB13730XL APPLIED BIOSYSTEMS sequence analyzer. The result of the sequence analysis was analyzed by BLASTN in the NCBI. The results were compared with data obtained from Gene Bank which are available at the NCBI online.
To determine whether the rs 4880 for SOD2 (T>C), rs4244285 for CYT2C19 (G>A), and rs1801274 for FCGR2A (T>C), were associated with susceptibility to breast cancer in the Iraqi women population living in the south of Iraq, we analyzed these SNPs in 25 healthy individuals and 50 patients with Breast cancer living in this region. Polymorphic analysis revealed that all three possible genotypes (TT, CT, and CC at the rs4880 site; GG, GA, AA at rs4244285; and TT, CT, and CC at rs 801274) could be detected for these SNPs. The TT genotype was the major genotype at the rs4880. GG genotype was the major genotype at the rs4244285 and TT genotype was the major genotype at the rs1801274 locus in the subjects studied.
The results revealed significant association between SOD2 rs4880 T, C-alleles and breast cancer where χ2 is 0.357 and P value equal 0.008, this indicates that there is a significant relationship between this snp and breast cancer. Also the results show that the frequencies were 56% for TT, 24% for CT and 20% for CC in G1 (Control). The frequencies were 24% for TT, 60% for CT and 16% for CC in G2 (Patients). There was significantly association in rs 4880 polymorphism of SOD2 between G1 healthy (control) and G2 patients (p<0.05) as show in Table 4. In G1, T allele frequency was 68% and the C allele was 32% and in G2 the frequencies were 54% and 46% for T and C alleles, respectively. Descriptive statistics analyses revealed that the rs4880 polymorphism, CT and CC genotypes were compared with the TT genotype there was significant (P= 0.01 <0.05), while when compared between CT and TT genotypes with CC genotype we found there was significantly difference. There is also no significant relationship between C allele with the reference T allele (p=0.101).
|
Polymorphisms SOD2 (C/T) |
G1 (Control) N=25(%) |
G2 (Pateints) N=50(%) |
χ2 |
P value |
OR (95%CI) |
P Value |
|
TT |
14(56.0) |
12(24.0 ) |
0.357 |
0.008 |
1.0ref (1.0ref) |
|
|
CT |
6(24.0) |
30(60.0) |
0.171(0.053-0.551) |
|||
|
CC |
5(20.0) |
8(16.0) |
0.536(0.138-2.082) |
0.002 |
||
|
0.365 |
||||||
|
T allele |
34(68.0) |
54(54.0) |
1.0ref (1.0ref) |
|||
|
C allele |
16(32.0) |
46(46.0) |
1.810 (0.888-3.691) |
|||
|
2.694 |
0.116 |
0.101 |
||||
|
TT |
14(56.0) |
12(24.0) |
1.0ref (1.0ref) |
|||
|
CT&CC |
11(44.0) |
38(76.0) |
0.248(0.089-0.690) |
|||
|
7.535 |
0.01 |
0.006 |
||||
|
CC |
5(20.0) |
8(16.0) |
1.0ref (1.0ref) |
|||
|
CT&TT |
20(80.0) |
42(84.0) |
0.762(0.221-2.627) |
|||
|
|
|
|
0.186 |
0.75 |
|
0.666 |
Table 4 The genotypes and allele distribution of SOD2 polymorphism in G1 and G2
The results revealed significant association between CYP2C19 rs4244285 A, G-alleles and breast cancer where χ2 is 0.295 and P value =0.032 <0.05, this indicates that there is a significant relationship between this SNP and breast cancer. Also the results show that the frequencies were 88% for GG, 12% for GA and 0% for AA in G1 (Control). The frequencies were 60% for GG, 32% for GA and 8% for GG in G2 (Patients). There was significantly association in rs4244285 polymorphism of CYP2C19 between G1 healthy (control) and G2 patients (p<0.05) as show in Table 5. In G1, G allele frequency was 94% and the A allele was 6% and in G2 the frequencies were 76% and 24% for G and A alleles, respectively. Descriptive statistics analyses revealed that the rs 424485 polymorphism, GA and AA genotypes were compared with the GG ( Dominant) genotype there was significant ( P= 0.013 <0.05 ), while when compared between GA and GG genotypes with AA( Recessive ) genotype we found there is no significantly association(P value =0.294)1. There is also significant association between G allele with the reference a allele (p=0.007). It was observed that the allele AA was not found in the G1 healthy, but was found to be 8% in patients with breast cancer. In addition, the ratio of allele A to allele G is very small in the G1 healthy while the percentage of allele A in patients was increased.
|
Polymorphisms CYP2C19 (G/A) |
G1 (Control) N=25(%) |
G2 (Pateints) N=50(%) |
χ2 |
P value |
OR (95%CI) |
P Value |
|
GG |
22(88.0) |
30(60.0 ) |
0.295 |
0.032 |
1.0ref (1.0ref) |
|
|
GA |
3(12.0) |
16(32.0) |
0.256(0.066-0.987) |
|||
|
AA |
0(00.0) |
4(8.0) |
1.733(1.373-2.188) |
0.038 |
||
|
0.095 |
||||||
|
G allele |
47(94.0) |
76(76.0) |
1.0ref (1.0ref) |
|||
|
A allele |
3(6.0) |
24(24.0) |
0.202 (0.058-0.708) |
|||
|
7.317 |
0.012 |
0.007 |
||||
|
GG |
22(88.0) |
30(60.0) |
1.0ref (1.0ref) |
|||
|
GA&AA |
3(12.0) |
20(40.0) |
0.205(0.054-0.775) |
|||
|
6.145 |
0.017 |
0.013 |
||||
|
AA |
0(00.0) |
4(8.0) |
1.0ref (1.0ref) |
|||
|
GA&GG |
25(100.0) |
46(92.0) |
1-543(1.300-1.832) |
|||
|
|
|
|
2.113 |
0.294 |
|
0.146 |
Table 5 The genotypes and allele distribution of CYP2C19 polymorphism in G1 and G2
The results revealed no significant association between FCGR2A rs1801274 T, C-alleles and breast cancer where χ2 is 0.097 and P value equal 0.735>0.05, Where it is noted that the P value is much greater than the 0.05 that there is no significant association between the healthy and patients with the alleles This indicates that there is no significant relationship between this snp and drug of breast cancer. Also the results show that the frequencies were 52% for TT, 32% for CT and 16% for CC in G1 (Control). The frequencies were 60% for TT, 30% for CT and 10% for CC in G2 (Patients). The percentage of alleles is close to the healthy and patients so there are no significant association as shown in Table 6. In G1, T allele frequency was 68% and the C allele was 32% and in G2 the frequencies were 75% and 25% for T and C alleles, respectively. Descriptive statistics analyses revealed that the rs 1801274 polymorphism, CT and CC genotypes were compared with the TT genotype there was no significant (P= 0.622>0.05), also when compared between CT and TT genotypes with CC genotype we found there was no significantly difference. There is also no significant association between C allele with the reference T allele (p=0438.).
|
Polymorphisms FCGR2A (T/C) |
G1 (Control) N=25(%) |
G2 (Pateints) N=50(%) |
χ2 |
P value |
OR (95%CI) |
P Value |
|
TT |
13(52.0) |
30(60.0 ) |
0.097 |
0.735 |
1.0ref (1.0ref) |
|
|
CT |
8(32.0) |
15(30.0) |
1.231(0.419-3.613) |
|||
|
CC |
4(16.0) |
5(10.0) |
1.846(0.426-8.006) |
0.705 |
||
|
0.409 |
||||||
|
T allele |
34(68.0) |
75(75.0) |
1.0ref (1.0ref) |
|||
|
C allele |
16(32.0) |
25(25.0) |
1.412(0.669-2.980) |
|||
|
0.822 |
0.438 |
0.364 |
||||
|
TT |
13(56.0) |
30(60.0) |
1.0ref (1.0ref) |
|||
|
CT&CC |
12(44.0) |
20(40.0) |
1.385(0.526-3.643) |
|||
|
0.436 |
0.622 |
0.509 |
||||
|
CC |
4(16.0) |
5(10.0) |
1.0ref (1.0ref) |
|||
|
CT&TT |
21(84.0) |
45(90.0) |
1-714(0.417-7.044) |
|||
|
|
|
|
0.568 |
0.709 |
|
0.451 |
Table 6 The genotypes and allele distribution of FCGR2A polymorphism in G1 and G2
Pharmacogenetic role of polymorphisms
From the pharmGKB database (Table 7), these three polymorphisms reported to be related to the efficacy or toxicity of breast cancer drugs, statistical analyses revealed that rs 4880 and rs 4244285 were significantly different in frequency in our samples and these polymorphisms related to cyclophosphamide and doxorubin. While rs1801274 related to Trastuzumab, doxorubicin, paclitaxel, cyclophosphamide. Because of Iraqi patients with breast cancer have significant association with rs 4880 and rs4244285 as we show in above statistical analysis and no significant association with rs1801274 so we suggest that drugs cyclophosphamide and doxorubin use as treatment to Iraqi patients with breast cancer. All these findings increase the overall knowledge on the prevalence of specific variants related with breast cancer treatment responsiveness in Iraqi population and highlight the importance of assessing gene polymorphisms with cancer medication in isolated communities.
|
Snp (gene) |
Drugs ( Reported as Related by PharmGKB Database) |
Alleles |
|
rs4880 |
Cyclophosphamide |
T/C |
|
(SOD2) |
||
|
rs 4244285 |
Cyclophosphamide |
A/G |
|
(CYP2C19) |
Doxorubicin |
|
|
rs 1801274 |
Trastuzumab,Doxorubicin |
T/C |
|
(FCGR2A) |
Paclitaxel, Cyclophosphamide |
|
Table 7 List of SNPs reported as associated with specific drugs in the PharmGKB database
Doxorubicine and cyclophosphamide are the backbone of the chemotherapeutic regimens used for the treatment of breast cancer patients. Cyclophosphamide is a pro-drug that needs to be oxidized to exert its cytotoxic effect. His step is catalyzed by a number of cytochrome P450 enzymes, including CYPC219.18 As regards CYP2C19, the rs4244285 variant has been reported to be associated with differential response to cyclophosphamide-doxorubicin adjuvant regimen. The frequency of the G allele in the Iraq cohorts. The GG genotype was the most frequent in Iraqi cohorts, individuals bearing the AA genotype show a trend of an increased risk of poorer outcome if treated with cyclophosphamide-doxorubicin polychemotherapeutic regimen.19 These results correspond to the results obtained by Massimiliano Cocca when studying this gene on the Italian population.20
Our study firstly suggests that SOD2 rs4880 and CYP2C19 rs4244285 polymorphisms play an important role in the development of breast cancer in Iraqi population, So this study has provided more evidence to support the concept that that the functional polymorphisms is significantly associated with BC. In addition that FCGR2A rs1801274 appear no significant association with BC in Iraqi women. Our explorative study highlights the importance of assessing gene polymorphisms related with cancer medications in isolated populations. In particular, the finding that specific functional variants, strongly associated with toxicity or lack of efficacy, are more prevalent in a specific community could lead to the development of regional targeted interventions aimed at a direct screening of such risk genes/variants for example by using targeted re-sequencing approach. His in turn could facilitate a more effective and rationale usage of the healthcare economic resources thus paving the way for a personalized medicine. Overall, rs4880 and rs4244285 appeared to influence the tolerance and effectiveness of (Cyclophosphomide and Doxorubicin) chemotherapy in this group of breast cancer patients.
There is no conflict of interest.
None.
None.

©2017 Jabir, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.