Journal of
eISSN: 2373-633X


Review Article Volume 3 Issue 1
University of South Florida, USA
Correspondence: Zheying Sun, University of South Florida, 2503 black horse loop, USA, Tel 8134548866
Received: May 24, 2015 | Published: August 13, 2015
Citation: Sun Z. The general information of the tumor suppressor gene p53 and the protein p53. J Cancer Prev Curr Res. 2015;3(1):208-217. DOI: 10.15406/jcpcr.2015.03.00068
This paper talks about the history of the human gene p53 and some important events since it's discovered in 1979. It also shows the detailed structure of the human gene p53 and the protein p53 which is encoded by the gene TP53. The proteins contain three major domain, N-terminal domain, DNA binding domain and C-terminal domain, each contains unique structure and function. Last, it describes the function of the protein p53 and talks about the future direction of the p53 study.
Keywords: the human gene p53, history, structure, function
EBNA-5: Epstein-barr virus nuclear antigen 5; HBX, hepatitis B virus X protein; NTD, N-terminal domain; CARD, caspase recruitment domain; AMPK, AMP-activated protein kinase; Rnai, RNA interference; Dox, doxycycline; LIF, leukaemia inhibitory factor; CTD, C-terminal domain; TD, tetramerization domain; RD, regulatory domain; NTD, N-terminal domain; DBD, DNA-binding domain; AD1, activation domain 1; VEGF, vascular endothelial growth factor
The gene p53 was first discovered in 1979. A protein was identified in simian virus 40-transformed mouse cells (SV40) by immunoprecipitation with anti-T serum; this protein was called protein p53.1 In the same year, Kress and other scientists found a new class of proteins with a molecular mass ranging from 50-60kDa. This kind of protein was then identified as p53.2 The protein p53 can also be identified from various transformed cell lines by immunoprecipitation. Lane and Linzer also got a similar result in 1979. Other evidence for identifying p53 is that p53 was expressed in all tested transformed mouse cells; these tests include chemically-induced sarcomas, transformed fibroblasts, and leukemias, while in normal cells, p53 was not expressed. Additionally, a high level of p53 was detected in most transformed cells no matter how the cells were transformed, either spontaneously or non-spontaneously.3 That was largely due to the increased stability of the p53, however, in F9 embryonic carcinoa cells, it expressed a high level of p53, this was due to the amount of translated p53 mRNA.4
After the protein p53 had been discovered in 1979, it became popular to analyze it. However, at that time, as it was a newly discovered protein, and there was not a former name for it, different institutions used different names and published papers with different names. In order to solve this problem, in 1983, during the 1st International p53 workshop held in Oxted, UK, scientists from different research groups in different countries got together to discuss a common nomenclature for this newly discovered protein. At this meeting, 'p53' become its name and it has been used since then. It was thought that the reason why scientists called the protein p53 is that the molecular mass of this protein is 53kDa that is based on its migration in SDS gel. Later the molecular mass was proved to be wrong, and the correct molecular mass should be 43.7kDa because p53 contains a proline-rich region, and this region can reduce the migration of p53 in SDS gel. But the name 'p53' remained.5
During the 1980s, the protein p53 was believed to be involved in the cell cycle, as well as playing a role in DNA replication. Later, in 1982 to 1994, people found that some viral oncoproteins were able to bind to p53, forming a complex. In 1982, Sarnow et al.,6 found that adenovirus E1b (58kDa) can interact with a 54kDa protein that is present in SV40-transformed mouse cells mentioned above. According to the results of immunological specificities of T antibodies and the peptide maps of the 54kDa protein, this 54kDa protein is identified as p53.6 In the same year, scientists found that if they injected the p53 antibody into Swiss 3T3 mouse cells it would inhibit cells entering the S phase of the cell cycle, however; under the same situation, the p53 antibody did not affect SV40 or adenovirus induced DNA synthesis.7
Later in 1984, scientists examined the effect of the p53 on non transformed 3T3 fibroblasts; they analyzed the synthesis rate of the protein p53 at different time points and found that in the late G1 phase, the synthesis rate and the level of the protein p53 and its related mRNA increase. This result suggests that the protein p53 inhibit cells entering the dividing phase from the interphase.8 Maltzman W et al.,9 did another experiment in the same year. They treated the non transformed mouse cell with UV light and UV-mimetic chemical carcinogen 4NQO, and they detected a high level of p53. The result showed that elevated expression of p53 is not only a symbol that indicates the cell cycle, but also more importantly a component that is involved in DNA synthesis and cell proliferation.9 In 1987, when studying the complex of T antigen of simian virus 40 and DNA polymerase α, Gannon, and other scientists found a similar change in the antigen when bound to p53 and polymerase α. They also found that at a certain concentration of the three components, they can form a special trimeric complex that includes T antigen, p53 and DNA polymerase α. As T antigen is involved in viral DNA replication and cellular transformation, this result indicates that p53 plays a role in the control of the cell cycle and DNA replication.10
As the experiment showed above, p53 has the ability to immortalize cells. In 1984, Eliyahu D et al.,11 found that p53 and the product of oncogene myc shared some similar properties. Both of them have the ability to bind to other proteins and are involved in the cell cycle, and they both accumulate in nuclei of transformed cells.11 Bienz, Pennica and Oren analyzed the amino acid sequences of the protein p53 and the product of myc, and they found that the two proteins show similarities in molecular structure and the position of special charged residues. Then scientists proposed that p53 may act as an oncogene. Based on this hypothesis, Eliyahu D et al.,11 did some experiments. As the primary rate embryo fibroblasts can be transformed by the involvement of both myc product and Ha-ras, primary baby rat kidney cells can also be transformed by cooperation of Ha-ras and adenovirus early region 1A,12 Eliyahu D et al.,11 decided to use this kind of biological test system to identify the oncogenic function of the p53. They treated normal embryonic cells with p53 and activated Ha-ras. The result showed that target cells encounter morphology changes and produce high levels of p53, Eliyahu D et al.,11] thought that the transformation of embryonic fibroblasts by p53 and Ha-ras explained that the gene p53 is an oncogene.11 In 1985, Jenkins proposed that the p53 gene can extend the lifespan of cells, enhance the affectivity of transformation by rearranging its coding sequence that could cause the production of stable proteins.13
However, in the late 1980s, scientists started to realize that p53 is a tumor suppressor gene instead of an oncogene. They observed that p53 with normal function cannot be detected in many of the tumors and found that losing the expression and function of wild-type p53 gene is necessary during cell transformation. These raise the possibility that wild-type p53 gene may inhibit neoplastic progression.14 Then they formulated another hypothesis: the clone gene p53 used in previous experiments contains dominant negative mutations within the highly conserved domain occasionally, which lead to opposite experiment results.15 In 1988, Ben and other scientists detected a huge amount of rearranged p53 in murine erythroleukemia cell lines--DP20-1 and CB3 which are derived from the spleens of murine infected with the Friend leukemia virus.16 In 1989, Eliyahu, who pointed out that p53 is an oncogene changed his mind, and he supposed that wild-type p53 gene may inhibit cell transformation. Eliyahu and other scientists studied the effect of wild-type p53 protein encoded by plasmids and mutant p53 on the ability to elicit primary rate embryo fibroblast transformation by various oncogene combination in vitro. For example mutant p53 plus ras, and myc plus ras. The result showed that wild-type p53 lead to a huge reduction of the transformed foci caused by mutant p53 plus ras; mutant p53 showed no inhibition on transformed foci caused by myc plus ras, while myc plus ras-mediated transformation is very sensitive to the expression of wild-type p53. Figure 1 shows this experiment concisely. It showed that compared with mutant p53, wild-type p53 exhibits an obviously inhibitory effect on the cell transformation. The effect is positively related to the expression level of wild-type p53 and negatively related to the expression level of mutant p53. This experiment suggested that wild-type p53 may indeed have an opposite function compared with mutant p53 and may inhibit the tumorigenesis.17 Currently, p53 is recognized as a tumor suppressor gene. It is estimated that about half of tumors are caused by p53. It is one of most frequently mutated genes in humans, and the most frequently analyzed gene around the world.5
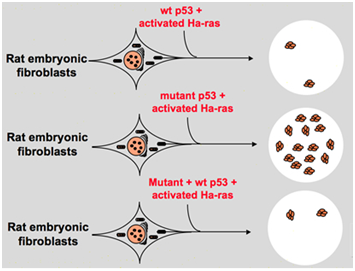
Figure 1 The experiment indicates that wild-type p53 is a tumor suppressor gene in vitro.17
During the first several years of the 1980s, the biochemical pathway of p53 and the effect of p53 mutation were not clear. In 1991, Kern and other scientists found that a 33-base pair DNA sequence binds specifically to wild-type p53 in vitro. They also found that the p53 protein contains two mutations that are usually found in human tumors that cannot bind to this specific DNA region. So they supposed that the function of p53 depends on its ability to bind specific DNA sequences, and this ability is altered by mutations found in human tumors. They also suppose that this 33-base pair DNA sequence may not be the only sequence that has the ability to bind specifically to the p53 in humans; however, it can help people better understand the function of p53.18 Later, p53 was found to play a role during the cell cycle, DNA repair, differentiation, initiating apoptosis and angiogenesis. Rotter V et al.,19 found that p53 up-regulates differentiation of cells. For example, a high level of p53 protein was detected in several key steps during B-cell differentiation. Elevated p53 can also be detected during spermatogenesis. Meanwhile, only a very low level of p53 protein can be detected in some organs of adult mice.20
In 1990, a useful tool was discovered occasionally. It is a temperature-sensitive mutant of p53, called p53val135. It can act as a real wild-type p53 at the temperature of 32.5oC, suppressing transformation, and it can also act as other mutated p53 at the temperature of 37.5oC or above 48oC, eliciting transformation. Additionally, for transformed cells expressing p53val135, its proliferation is controlled at the permissive temperature, and this kind of control is reversible. By using this p53val135 mutant, wild-type p53 was discovered to induce cell cycle arrest at either G1 or G2/M.19 In 1991, Elisheva et al.,21 found that the temperature-sensitive p53val135 performed a different function in the murine myeloid leukaemia cell line. After reactivation of p53val135 for a few days, all cells died, and this death exhibits some properties of apoptosis.21 A year later, a similar result was obtained by Shaw. A wild type p53 was transfected into a human colon tumor-derived cell line EB. The cells were examined under light and electron microscopes, and found to exhibit some properties of apoptosis.21 In 1990, Scheffner et al.,22 and other scientists found that E6 that stimulates the destruction of host cell regulatory proteins is encoded by the oncogenic human papillomavirus types 16 and 18, and it can form a complex with wild-type p53 in vitro, which in turn causes the degradation of protein p53.23
In 1992, a key protein MDM2 was discovered because it binds tightly with p53, and it inhibits the transactivation mediated by p53. The molecular mass of MDM2 is 90kDa, and it forms a complex with both mutated and wild-type p53.24 In the same year, Livingstone RL et al.,25 studied whether the cell lost one or both copies of wild type p53 alleles and if that was sufficient to cause gene amplification. Gene amplification was detected mostly in transformed cells but not in the normal fibroblasts. The result showed that cells losing one copy of the p53 alleles act as wild-type p53, while cells losing both copies of the wild type p53 alleles exhibit a higher frequency of amplification.26 Another experiment made by Yin Y et al.,27 showed a similar result.25
In 1993, a p53 target gene called CDKN1A was identified. It encodes the protein p21 which is a cyclin-dependent kinase inhibitor that inhibits cyclin-CDK2 and CDK1 by binding to them. In 1993, Szekely found that the Epstein-Barr virus nuclear antigen 5(EBNA-5) is encoded by the Epstein-Barr virus, and it can infect human B lymphoblastoid cell. A 66 amino acid long peptide is responsible for the formation of complex EBNA-5-p53, point mutations of p53 did not affect its binding ability to EBNA-5. However, it inhibits its formations of complexes with other molecules.27 In 1994, Cho and his co-workers first described the crystal structure of complex p53-DNA. This DNA binding domain was also called the core domain. It contains residues 102-292, and consists of a beta sandwich. They also demonstrated the detailed structure of the core domain.28 Also in 1994, Wang XW et al.,29 the interaction between hepatitis B virus X protein (HBX) and wild-type p53 protein in humans. They found that HBX can inhibit the ability of p53 to bind to other sequence-specific DNA after it is bound to p53 and it can also inhibit the association of p53 with transcription factors.30
In 1997, Honda R et al.,31 first hypothesized that MDM2 can trigger p53 ubiquitylation and lead to degradation of p53 by a ubiquitin-proteasome system. They pointed out that MDM2 binds to the N-terminal domain (NTD) of p53 and acts as ubiquitin ligase E3.29 Also in 1997, two new families of proteins, p63 and p73 were discovered that share substantial homology with p53. p73, also called tumor protein 73, is encoded by a gene located in 1p36. The location is deleted frequently in neuroblastoma and other tumors. p73 can activate p53 target genes and interacts with p53.31 Yang et al.,32 found that the gene p63 is located in 3q27-29 and it can be detected in various mouse and human cells. Like p73, p63 can transactivate p53 target genes significantly, it can also induce apoptosis. One characteristic of p63 is that the majority of p63 lack an N-terminus.32 In the same year, Serrano and co-workers found that primary murine fibroblasts can be transformed by ras in the absence of p53 or p16, and inactive p53 or p16 can facilitate the immortalization process of human cells. These findings suggest that p53 plays a role in cellular senescence.33 Then, in 1997, p53 was found to play a role in the initiation of apoptosis. When cells come into the proliferation phase, the telomeres at the end of each chromosome would shorten after each round of DNA replication due to incomplete replication of single strand DNA at the end of the DNA stand.34 Activated tumor suppressor gene p53 limits the number of times cell division can occur. Wynford TD35 found that with the loss of the function of wild-type p53, all fibroblasts escape from apoptosis. Also, the transactivation function of p53 can be turned on by apoptosis.36 Wynford TD35 proposed that there are three possibilities of how p53 is activated. The first one is post-translational modification by phosphorylation, the second one is up-regulates the transcritional cofactors like p33ING1, the last one is down-regulates the p53 inhibitors like MDM2.36
In 2000, Brodsky MH et al.,37 studied the transcription targets of p53 in Drosophila. There is evidence to show Drosophila eyes display a severe rough eye phenotype under the expression of human p53 that will induce apoptosis of eye imaginal disc cells, causing the loss of pigment cells, finally inhibiting eye development of Drosophila,35 so Drosophila can be a model animal for studying the function of p53. Brodsky found that the gene rpr contains a consensus p53 binding site that is located in the cis-regulatory region of rpr, and it also is an activator of apoptosis. With other evidence, Brodsky claimed that rpr is one transcriptional target of the p53.38 In 2001, Derry and co-workers found that C. elegans do not have a p53 gene, but indeed contain a gene cep-1 that encodes proteins that have a similar sequence with protein p53. This C. elegans gene encodes protein CEP-1 which has the ability to induce apoptosis by genotoxic stress and is a necessary component during meiosis.39
In 2002, Tyner and co-workers proposed that p53 plays a role in regulating the aging of organisms. In order to study the function of p53, they created genetically engineered mice with mutated p53 by deleting exons 1-6 and an upstream region of wild-type p53 gene (p53+/+), called p53+/m. It act as wild-type p53 and has enhanced resistance to spontaneous tumors better than wild type p53. In the experiment, they monitored the mice containing p53+/m, p53+/+ and p53+/-. p53+/- means the mice lossing one copy of the wild-type p53 gene. The results showed that none of the mice with p53+/m developed life-threatening tumors, however, more than 80% of mice with p53+/- and more than 45% of mice with p53+/+ developed these kinds of tumors. Looking inside of the tumors, localized tumor lesions were observed in 2 out of 35 p53+/m mice, in contrast, various tumors like lymphomas, and osteosarcomas were found in p53+/- and p53+/+ mice. During this experiment, they also observed that the median age of p53+/m was 96 weeks while the median age of p53+/m was 116 to 118 weeks. Tyner and co-workers also examined the possibility that the shorter life of p53+/m was associated with aging. They found that after 18 months, the p53+/m mice began to lose weight and vigour, as for p53+/m mice, reduced weights were observed at age 30-36 months. p53+/m mice also exhibit lordokyphosis. Depending on X-ray analysis, p53+/m mice exhibited reduced bone density in the age of 12 months, and it will become severe at the age of 18 months. This is a symbol of osteoporosis and osteoporosis is a marker of aging in humans and mice.40 Tyner et al.,41 also tested the tolerance of stress, as this ability is also a marker of aging.42 They performed 3 mm punch biopsies in the back skin of old and young anaesthetized p53+/m and p53+/+ mice. Their results showed that a lot of old p53+/m mice died after injected standard dose of Avertin, indicating that old p53+/m mice were less tolerant to the stress.43
In 1991, it was found that p53 has the ability to induce apoptosis, while in 2003; Mihara and other scientists found that p53 also has an apoptosis role in the mitochondria.41 Since some mitochondrial proteins have the ability of activating cellular apoptosis either by active caspases or neutralizing cytosolic inhibitors. In the example of cytochrome c-induced caspase, after receiving the apoptosis signal, cytochrome c is released from the intermembrane space of the mitochondria, and then in turn binds to Apf-1 which exists as an inactive monomer, induces its conformational change, and increases its binding affinity for dATP/ATP by 10-fold than Apaf-1 binds dATP/ATP alone. Then the complex Apaf-1- cytochrome c binds to dATP/ATP, form the apoptosome. After that the caspase recruitment domain (CARD) of Apaf-1 exposed in the apoptosome, recruit procaspase-9, and then autoactivate themselves. The final complex then cleaves and activates other caspases such as caspase-3 which in turn subsequently cleave important molecules in the cell, causing chromatin condensation, DNA fragmentation and finally leading to apoptosis.44 Figure 2 shows the cytochrome c-induced caspase activation pathway.
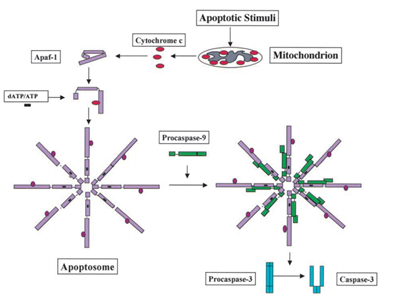
Figure 2 The cytochrome c-induced caspase activation pathway.44
Mihara M et al.,44 scientists found that the wild-type p53 gene can be translocated to the mitochondrial surface of tumor cells rapidly. In the experiment, they found that some stress-induced wild-type p53 protein has the ability to translocate to the mitochondria of thymocytes in human or mouse cells after apoptosis due to DNA damage and hypoxia. Then these wild-type p53 proteins induce permeabilization of mitochondria and cause a series of changes that occur in mitochondria like releasing cytochrome c by forming complex with Bcl2 and BclXL.44
As a good clinical result with little side effect, gene therapy is popular. By the end of 2005, there were 1020 gene therapy trials in the database of Journal of Gene Medicine. Among these trials, 66% of gene therapies were conducted on cancer patients, and 58 trials of this used rAd-p53, a recombinant adenovirus encoding the human p53 gene. In April 2004, a recombinant human adenovirus-p53 injection (Gendicine) was launched formally. Gendicine is used to treat head and neck squamous cell carcinoma and it was approved by the State Food and Drug Administration of China on Oct. 16, 2003. It became the first gene therapy product in the world to be approved by the Chinese government.45
The gene p53 was discovered to regulate metabolism in 2005. In order to transfer from G1 to S phase, cells must have sufficient raw materials support for DNA, organelles and protein synthesis. To regulate this process, some checkpoints are necessary. One of them is the glucose-dependent checkpoint at G1/S. It is regulated by the AMP-activated protein kinase (AMPK). When glucose is exhausted, AMPK can phosphorylate protein p53, which in turn induces cell arrest, and avoids cell death. Cells which encounter the p53-dependent arrest will reenter the cell cycle when glucose is sufficient.46
It is known that inactivation of p53 is necessary for the formation of tumors. Bykov et al. VJ and Snydel EL et al.,47–49 point out that improper functioning of p53 can lead to the proliferation of an existing tumor.47,48 Ventura and his co-workers did some experiments to test this hypothesis. They restored the function of endogenous p53 in primary autochthonous tumors to examine the consequence of p53 reactivation. The result showed that p53 reactivation was responsible for regression of autochthonous tumors. That means inactivated p53 protein can lead to tumor development.49 Xue and other scientists also did an experiment to test the consequence of reactivating p53 on tumors. They used reversible RNA interference (RNAi) to regulate the expression of endogenous p53 in mice with liver cancer. In the experiment, doxycycline (Dox) is used to reactivate p53, as the expression of p53 is totally suppressed when Dox is missing and rapidly restored when Dox is added. When treated with Dox, p53 miRNA was shut off which in turn causes increased expression of p53. The result showed that the tumors in Dox-treated mice become undetectable after 12 days, while tumors in untreated mice grew rapidly. To test the consequence of transient reactivation of p53, they treated mice with Dox for 4 days and then stopped. The result showed that even a two-day treatment can cause regression of tumors and 4 days of treatment can cause the tumors to completely regress. They also pointed out that during tumor regression, transiently-reactivated p53 can trigger cellular senescence, not apoptosis. In the same year, Hu found that embryonic implantation in p53-/- female mice is regulated by the Leukaemia inhibitory factor (LIF). The LIF is a secreted cytokine and is important for blastocyst implantation. The gene coding LIF is identified as the p53 target gene and the p53 binding site is located in intron 1 in both humans and mice.50,51
Basic information on the human gene p53
The protein p53 is encoded by the TP53 gene in humans. TP53 spans about 20 kb and is located on the p arm of the chromosome 17 (17p13). Figure 3 shows the location of the p53 gene family (p53, p63, p73) and other regulator genes in human chromosome 17. In 1985, McBride, Merry, and Givol used the cloned human p53 cDNA as a probe to examine the location of p53. Then, using in situ hybridization, they confirmed that p53 was located on the most distal band of the short arm of chromosome 17.52 In situ hybridization is a type of hybridization that uses a labeled DNA or RNA probe to localize a specific DNA or RNA sequence in a part of tissue or cells. Figure 4 shows that the gene TP53 contains 11 exons in which the first one is a non-coding exon and is 8-10 kb away from exon 2. Figure 4 shows the structure of the human p53 gene.53
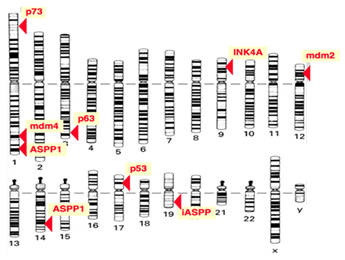
Figure 3 Location of gene TP53.52
Transcription of the p53 gene can be initiated from two different sites: P1 and P1' is located at the upstream of exon 1, and P2 is located in intron 4. The transcript from P2 will synthesize a protein lacking an N-terminal. α, β and γ shown in Figure 4 are locations for alternative splicing, resulting in three isoforms: P53, P53β, P53γ. Proteins that are encoded by P53β and P53γ lack the oligomerization domain. Therefore, the human p53 gene can encode at least nine different p53 proteins and fifteen different mRNA due to alternative splicing locations and alternative promoters.54 The genes p63 and p73 were identified in 1997. They have similar DNA binding domain with gene p53, and thus share the same properties that can cause apoptosis and cell cycle arrest. However, transgenic knockout mice that contain one of the p53, p63 and p73 exhibit distinct phenotypes, indicating that each of them contain specific functions. Figure 5 shows the human p63 and p73 gene structures. In the gene p63, P1 and P2 are alternative promoters. In this gene, α, β and γ shown in Figure 5 are locations for alternative splicing. TAp63 and ΔNp63 are p63 protein isoforms when the transcript is initiated at P1 and P2. In the gene p73, P1 and P2 are alternative promoters. In this gene, α, β, γ, ζ, Δ and ε are locations for alternative splicing. TAp73, Ex2p73, Ex2/3p73, ΔN'p73 and ΔNp73 are p73 protein isoforms due to alternative splicing and promoters.
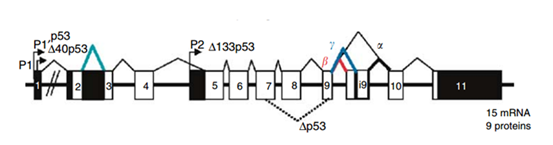
Figure 4 Human p53 gene Structure.53
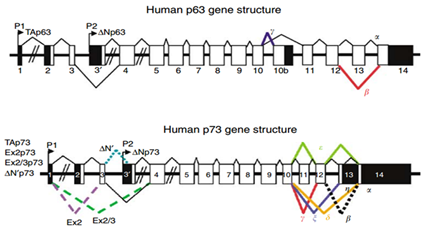
Figure 5 The schema of human p53 gene and p73 gene structure.54
Basic information on the human protein p53
Most diseases are related to incorrect expression of the gene p53 and the dysfunction of the p53 protein. The protein p53 contains three domains. Figure 6 shows the structure of the human protein p53 and its domains. In this figure, it is clearly shown that the human p53 protein contains three major domains: N-terminal domain (NTD), DNA-binding domain (DBD), and C-terminal domain (CTD). NTD consists of the acidic N-terminus transcription activation region and a proline-rich region. CTD consists of a tetramerization domain (TD) and a regulatory domain (RD)
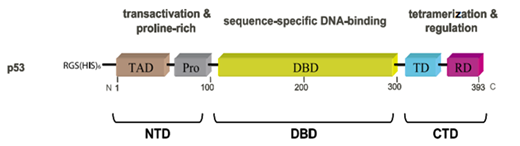
Figure 6 p53 protein structure and its fragments.55
Figure 7 shows more details about the protein p53 and other factors. The top of the figure shows proteins and viruses that can interact with the p53 protein. The N-terminal region can recruit some transcription factors like TBP, TFIID and so on. It can also interacts with RP-A that can bind to single strand DNA. MDM2 binds this region and negatively regulates p53 in two ways. The first one is MDM2 bind to p53 and inhibit the transcription of p53 directly. Secondly, MDM2 would stimulate p53 degradation rapidly. The central region contains four conserved regions, Ⅱ, Ⅲ, Ⅳ, Ⅴ shown in the third line. Zn also exists in this domain. About 80% p53 mutations related to cancer are within this region. The C-terminal region contains TD and RD. The structure of TD was solved in 1994 by X-ray crystallography and NMR.
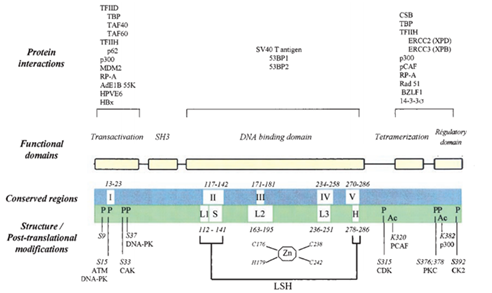
Figure 7 Schema structure of p53 protein and other related molecules. The third line shows structures and post-translational modification. "L" means loop and "LSH" means a loop-sheet-helix motif that was first described by Cho. "P" and "Ac" means phosphorylation and acetylation.56
The N-terminal domain (NTD) of the p53 Protein
The first one is N-terminal domain called NTD. It contains two transcriptional activation domains, which are residues 1-42 and residues 55-75. The major one is an acidic N-terminus transcription activation domain (TAD), also called activation domain 1 (AD1). It consists of the amphipathic helix and is used to activate transcription factors and plays an important role in the regulation of several pro-apoptotic genes. The acidic amino acid sequence located at the N-terminal has a similar function with transcription factors. After it combined with GAL4 whose product is a positive regulator of expression of galactose-induced genes, the recombinant protein initiate the transcription of the GAL4 operon. Activation domain 2 (AD2) is comprised of residues 43-64, and it is important for apoptotic activity. The entire TAD amino acid contains 73 residues, and it has a charge of -17 because of its acidic amino acid rich sequences. The acidic amino acid in the N-terminal makes it form an α helix easily. The transcriptional activity of TAD is higher than the sum of activities of AD1 and AD2.55 Also, a nine-amino-acid trans activation domain (9aaTAD) was found in domain AD1 and AD2.
Residues 64-92 is the proline-rich region, and it is important for apoptotic activity of p53. Venot and his co-workers studied the function of the proline-rich domain. This domain contains five repeat motif PXXP, which can bind to SH3. They produce a proline missing p53 (Δ pro) by a PCR-based method. The results showed that Δ pro lacking proline-rich domain slows down the expression of promoters like MDM2 and WAF1. This means that this proline-rich domain is necessary for p53 activity which includes apoptosis, reactive oxygen species and other function of p53.56
Well and his co-workers identified the structure of an intrinsically disordered region in TAD domain by residual dipolar coupling (RDC). They can measure RDCs by NMR spectroscopy and small-angle X-ray scattering (SAXS) not only in isolated TAD fragments, but also in intact p53 protein.57 Iakoucheva et al.,58 found that some proteins lack the intrinsic 3D structure when they perform their function. More than 30% of eukaryotic genes are able to code intrinsically disordered proteins that are longer than 30 amino acids, and 80% of intrinsically disordered proteins are associated with cancer. Moreover the disorder domain maybe involved in transcription, translation, DNA replication and DNA repair. So it is important to study these kinds of protein. Understanding the properties and functions of intrinsically disordered proteins is important to study unstructured regions in the human p53 protein.58
The intrinsically disordered protein is a protein that lacks three-dimensional structure. Most of them contain disordered linker regions between two global or transmembrane domains, and can be transformed to an ordered confirmation when they bind to their target molecules. A few intrinsically disordered proteins do not undergo confirmation changes; they maintain disordered structures and the structure disorder in complex with other molecules can be either static or dynamic. The structure of intrinsically disordered protein can be identified by experimental methods like NMR spectroscopy and X-ray crystallographic. It can also be identified by the computational method. Some software’s are used to predict disordered regions by exploring protein sequences, like IUPRED, Dispopred.
About 50% of p53 consists of an intrinsically disordered region. Scientists reviewed more than 90 proteins and found that most of the disordered regions in protein were involved in the cell cycle or regulation by interacting with DNA, RNA and other molecules in cells. When proteins bound to target molecules like DNA, their unstructured regions refolded and in turn changed the conformation. Dawson and other scientists also examined the interaction of Np53 and Mdm2, a negative regulator of p53 gene, by fluorescence spectroscopy. The result showed that NTD domains functioned synergistically with other folded domain in p53 when interacting with Mdm2.59 Vise and other scientists studied the reaction between p53 and the human replication protein A (hRPA70), a DNA binding protein that is required for DNA repair, by nuclear magnetic resonance spectroscopy. The result showed that p53 residues 40-60 located in the TAD region are necessary for interaction with hRPA70.60
Lum and other scientists studied the response of four segments in p53 TAD when binding to the N-terminal domain of Mdm2. They found that the confirmation of TAD in p53 changes from random coil after binding Mdm2 by fluorescence quenching and fluorescence correlation spectroscopy. The result is the same as those derived from nuclear magnetic resonance spectroscopy.61
Instead of studying the structural organization of intact p53, Dawson and co-workers studied the structure and function of the isolated N-terminal domain of p53 by examining the physical and structural parameters using the biochemical and biophysical methods. To analyze the secondary and 3D structure of isolated N-terminal region of p53 (Np53), they recorded the CD spectra of isolated Np53 in the far-UV region. The result showed that the minimal value of the CD spectrum of Np53 fragment is 200nm; this means a lot of α-helices andβ-sheets are missing. In the control experiment, they record the CD spectra of denatured Np53 in the far-UV region, the result shows little difference with that under native conditions. To analyze the quaternary structure and hydrodynamic parameters of isolated Np53, they used analytical size-exclusion chromatography and ultracentrifugation. Their results showed that Np53 molecular mass gained from the experiment is 23kDa and 24kDa, which is much higher than the theoretical molecular mass (Np53 migrated in SDS-PAGE shows a molecular mass of 23kDa, Np53 eluted from the gel-filtration column shows a molecular mass of 24KDa). This evidence suggests that the isolated Np53 fragment is an unfolded region under a native station.59
The DNA-binding domain (DBD) of the p53 Protein
The second domain contains residues 102-292. It is a central DNA-binding core domain, also called DBD. It contains one zinc atom and several Arginine amino acids and it is used to bind the p53 co-repressor LM03. Mutations of p53 usually occur in the DBD region and most commonly at sites 175, 248, 249, 273 and 282.62 Bargonetti et al.,63 used DNase I footprint assay to study the interactions between the wild-type p53 protein and the mutant p53 protein with SV40 DNA. They found that wild-type p53 binds specifically to DNA sequences, while mutant p53 did not bind specifically to these sequences.63 Then Vogelstein b et al.,64 proposed that p53 performed its function by binding to specific DNA conscious sequence in 1992, and after analyzing a large number of human genomic cDNA which can interact with p53, they found two copies of a 10 bp motif exist in each cDNA. The two motifs have symmetrical structures, each containing half of the sequence and form head-to-head quarter sites.64 Also Funk WD et al.,65 and other scientists found that p53 can initiate the transcription of the genes that contain p53 binding sites. All this evidence shows that there is a relationship between normally functioning p53 and DNA binding sites.65
In order to identify the location of the DNA binding site of p53, Pavletich and co-workers used a simple method, proteolytic digestion by subtilisin, to localize DNA binding domain in p53 protein. After digestion of the p53 protein, they injected fragments of p53 into E.coli and expressed them in vitro. The results showed that the terminal region is easily digested and folded loosely, the middle region is hard to digest and folded compactly, and the core domain contains DNA-binding site.62 Wang studied the p53 domain and found that residues 80-290 bind DNA either specifically or nonspecifically, and they do not form tetramers, while residues 280-290 binds DNA nonspecifically and form tetramers.66
The C-terminal domain (CTD) of the p53 protein
The third domain is the C-terminal domain, also called CTD. Residues 316-325 serve as the nuclear localization signaling domain. Residues 307-355 are the homo-oligomerisation domain (OD), which is also called the tetramerization domain (TD). The structure of oligomerisation domain was confirmed by NMR and X-ray. The oligomerisation domain consists of two symmetric dimers. Each dimer contains two antiparallel α helices and one antiparallel β sheet. The dimers are necessary for the activity of p53 because mutated p53 that lack oligomerization fail to suppress carcinoma cell.67 Residues 356-393 form a regulatory domain (RD) which down-regulates DNA binding of the central domain.
Wild-type p53 contains unstructured regions besides the folded regions mentioned above. Bell and other scientists point out that wild-type p53 contains a portion of unstructured region in its N-terminal and C-terminal by CD spectra analyze.68 As a result, p53 is also called an intrinsically disordered protein (IDP).
The protein p53 is related to cell senescence
Recent studies show that senescence in humans is caused by shorter telomeres.49 Compared with apoptosis, senecence just inhibits the proliferation of tumor cells, and cannot eliminate them from tissues. Cells during senescence undergo morphological changes, like cellular enlargement, and increased synthesis of lysosomes. Scientists found that after each round of DNA replication, 50-200 bp of telomeric DNA remains unpaired, and these unpaired DNA can be eliminated. They also found that human cells will undergo senescence when the length of telomeres is reduced to 4-7 kb. Early in 1997, Serrano and co-workers found that oncogenic ras induce cells senescence associated with evaluated p53 and p16INK4a. They injected the ras allele, Ha-rasV12 to 3 cell lines: primary human diploid fibroblasts (IMR 90 and WI38), primary mouse embryo fibrobasts (MEFs) and the rat cell line (REF52). The results showed that all cells exhibit 5 to 10 fold increases in Ras protein and all cells undergo morphological changes which is similar to that in senescence. They also observed increased level of p53, p16 and decreased levels of cyclin A and CDK2 kinase activity during this process. CDK2 is an enzyme which is essential for the transition of cells from G1 to S phase.34 Other scientists also found that over expression of some tumor suppressor genes like p16 cyclin-dependent kinase inhibitor (CDKI) and promyelocytic leukemia protein (PML) will strongly induce cellular senescence. Scientists proposed that p53 was related to cell senescence, because they found that the level of p53 is elevated transiently and then dropped to the normal level during some cell senescence. They also found that the DNA binding activity and transcription activity of p53 are increased.69
The protein p53 can regulate the cell cycle
The protein p53 can inhibit transformations of cells because it cannot tolerate any abnormal situations and stimuli. When DNA is damaged, cell division arrest at phase G1/S, G2/M in the majority of organisms. Checkpoint G1/S can inhibit the replication of damaged template DNA, offer the opportunity for damaged DNA to repair, extend survival time of damaged cell, and inhibit the proliferation of cells that contain damaged DNA. Early in 1992, Livingstone and co-workers found that the lack of wild type p53 can alter cell cycle arrest and gene amplification by changing the cell cycle progression.26 In 1998, Ikeguchi and other scientists found that if the DNA damage occurs in early G1, p53 can trigger checkpoints, and prevent damaged cells from entering the S phase. P21WAF1/CIP1 is required for this process. The protein p53 can binds the promoter sequence of P21WAF1/CIP1, initiates the transcription of P21WAF1/CIP1, and promotes the synthesis of protein P21WAF1/CIP1. The protein P21WAF1/CIP1 would inhibits the function of complex CDK-cyclin and cell grow, as well as inhibits the activity of PCNA after binding it. It can also control the S phase of the cell cycle by preventing DNA replication. Protein P21WAF1/CIP1 can also induce G1 arrest by preventing the phosphorylation of pRb which is a substrate for CDK2. The protein P21WAF1/CIP1 can also prevent cells from entering the mitotic phase by combining with cyclin A and B.70 Once DNA damage has been repaired, MDM2 allows the cell to reenter the cell cycle. p53 induces the expression of MDM2, then it binds to MDM2 which in turn deactives itself, and allows cells to pass the checkpoint and enter S phase.71
The protein p53 can trigger cell apoptosis
Apoptosis is another cell response for DNA damage. p53 regulates some apoptosis-related genes. The pro-apoptotic genes include Bax, Bak, Noxa and other genes, the anti-apoptotic genes include Bcl-2, Bcl-XL, Bcl-B and other genes. Bcl-2 is a key regulator during tumorigenesis and apoptosis. Bcl-2 and Bax are homologous proteins. Studies show that Bcl-2 inhibits most apoptosis pathways while Bax accelerates apoptosis. Bcl-2 can form heterodimers with Bax in cells, Bax can form homodimers with itself. Apoptosis depends on the ration of heterodimers and homodimers in the cell. The promoter of Bax contains a p53 binding site; p53 can promote transcription of Bax directly. Another factor IGF is an against apoptosis factors that in turn avoids cell death. The gene p53 can induce cell apoptosis by regulating IGF. It can also induce cell apoptosis by interfering transduction pathway of growth factor.72 Figure 8 shows the cell apoptosis pathway mediated by death receptors and mitochondria. In the death receptor pathway, death ligands bind to extracellular death receptors, forming death-inducible signaling complex (DISC), then DISC activates caspase 8 with an adaptor protein, such as FADD and procaspase 8. The activated caspase 8 then cleaves the inactive precursor enzyme procaspase 3, forming activated caspase 3 that induces apoptosis. The apoptosis pathway is mediated by mitochondria; members of Bcl-2 family are involved in it. As mentioned above, p53 causes the outer membrane of mitochondria to permeabilise, resulting in the release of cytochrome c, thus forming an apotosome with Apaf 1 and procaspase 9. Then the apotosome will activates caspase 9 and in turn activates caspase 3. Smac/DIABLO and Omi inactive IAPs, the inhibitor of apoptosis protein. Apopotosis-inducing factor (AIF) and EndoG promote chromatin fracture. p53 activates pro-apoptotic genes and inactivates anti-apoptotic genes during this process.73
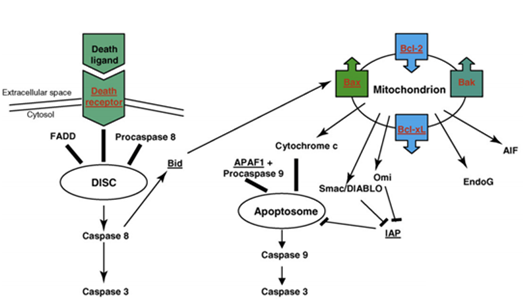
Figure 8 Schematic of apoptosis pathway mediated by death receptor and mitochondrial.78
The protein p53 can regulate angiogenesis
Angiogenesis is essential for the process of normal cells transformed to tumors. Thrombospondin-1 (TSP-1) is a potent inhibitor for angiogenesis. p53 stimulates the endogenous TSP-1 gene, and positively regulates the promoter sequence of TSP-1. Studies show that wild-type p53 can inhibit angiogenesis by regulating TSP-1. Another study found a significant correlation between the p53 and the vascular endothelial growth factor (VEGF). As the mutation rate of p53 increases, the number of capillaries increase. This suggests that the p53 mutation up regulates VEGF, which in turn promotes angiogenesis. All this evidence shows that wild-type p53 gene can inhibit angiogenesis.74 In contrast, Bernard and co-workers found that Δ133p53α and Δ133p53γ stimulate angiogensis in human glioblastoma U87.75,76
The relationship between the mutated p53 gene and cancer
As mentioned earlier, 50% of cancers are related to the mutated p53 and 80% of mutations are located in the DNA binding domains of the protein p53. Scientists found that mutated p53 gene was involved in breast cancer, lung cancer, and other tumors directly.
The future direction of The Protein p53 study
As we already known that the protein p53 is an effective target for cancer treatment ranging from traditional radiation to novel therapies. And scientists will continuously study this molecule in the fields of toxicology, pharmacology, and gene therapy. Scientists already found that how far the drugs that target p53 have progressed down the clinical pipeline.77 Currently, there are several methods to treat cancer through p53 protein pathway. These methods include reactive the wild-type p53 gene, inactive the ubiquitin ligase and p53 target genes that down regulate the p53, using alternative molecules that have the similar function with p53.78
During these process, scientists need to know the efficiency of the inactivation. Thus the methods that measure the gene expression are important. Gene expression can be quantified by measuring either mRNA or the related protein. Now, the most efficient and popular way is DNA microarray. It is a technique to query the expression of hundreds to thousands of genes simultaneously. It will save a lot of time and money during experiments.
One future direction of p53 study is bioinformatics and biostatistics. Both of them are using mathematics and software to analyze the biological data. It allows people freed from the boring experiments and get the credible results. Another future direction of p53 study is proteomics. It is a kind of method that large-scale analysis of proteins expressed in a given biological system. By analyzing the data, we may get the rough protein structure.
Professor El-Rady and Te-Strake helps me a lot when I write this paper. Te-strake helps me define my topic and narrow it, El-Rady direct my writing and correct the paper content.
The authors have no financial conflicts of interest to declare.
None.

©2015 Sun. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.