Journal of
eISSN: 2469 - 2786


Review Article Volume 6 Issue 1
Department of Biology, State University of New York, USA
Correspondence: Hon H Ho, Professor of Biology, State University of New York, New Paltz, NY 12561, USA
Received: January 23, 2018 | Published: February 12, 2018
Citation: Ho HH. The taxonomy and biology of Phytophthora and Pythium. J Bacteriol Mycol Open Access. 2018;6(1):40-45. DOI: 10.15406/jbmoa.2018.06.00174
The genera Phytophthora and Pythium include many economically important species which have been placed in Kingdom Chromista or Kingdom Straminipila, distinct from Kingdom Fungi. Their taxonomic problems, basic biology and economic importance have been reviewed. Morphologically, both genera are very similar in having coenocytic, hyaline and freely branching mycelia, oogonia with usually single oospores but the definitive differentiation between them lies in the mode of zoospore differentiation and discharge. In Phytophthora, the zoospores are differentiated within the sporangium proper and when mature, released in an evanescent vesicle at the sporangial apex, whereas in Pythium, the protoplast of a sporangium is transferred usually through an exit tube to a thin vesicle outside the sporangium where zoospores are differentiated and released upon the rupture of the vesicle. Many species of Phytophthora are destructive pathogens of especially dicotyledonous woody trees, shrubs and herbaceous plants whereas Pythium species attacked primarily monocotyledonous herbaceous plants, whereas some cause diseases in fishes, red algae and mammals including humans. However, several mycoparasitic and entomopathogenic species of Pythium have been utilized respectively, to successfully control other plant pathogenic fungi and harmful insects including mosquitoes while the others utilized to produce valuable chemicals for pharmacy and food industry.
Keywords: oomycota, pythiaceae, peronosporaceae, straminoplia, chromista
Members of the genera Phytophthora de Bary and Pythium Pringsheim are economically important “fungal-like-organisms” or “pseudofungi” which have been widely studied throughout the world but their taxonomic treatment has been highly controversial. The production of globose oogonia in the sexual reproduction has placed them in the phylum Oomycota. Although members of Oomycota are similar to all filamentous fungi they have been excluded from Kingdom Fungi and placed in the Kingdom Chromista1 along with several photosynthetic algal groups: brown algae (Phaeophyta), yellow green algae (Xanthophyta), golden algae (Chrysophyta) and diatoms (Bacillariophyta), that produce heterokont zoospores with two laterally inserted flagella, the anterior being tinsel-like and the posterior being smooth and whiplash, inserted at the same point. In order to better distinguish the colorless oomycetes from the pigmented algal allies, the term “straminenopile” was introduced by Patterson,2 based on the tubular hairs (mastigonemes) on the anterior flagellum of a heterokont zoospore as “straw with hairs”. It was formalized as a new kingdom Straminipila by Dick.3 The diploid (2N), cellulosic mycelium of oomycetes differs from the true fungi in producing oospores in sexual production and heterokont zoospores in asexual reproduction, whereas true fungi have chitinase, haploid (N) or dikaryotic (N+N) mycelium producing zygote, zygospores, ascospores or basidiospores in sexual reproduction and no zoospore or zoospore with single posterior whiplash flagellum in asexual reproduction.
The exclusion of oomycetes from Kingdom Fungi has been supported by molecular biology studies which however presented new problems to the classification of Phytophthora and Pythium. Morphologically, members of both genera are very similar in having coenocytic, hyaline and freely branching mycelium although the hyphae of Phytophthora species are usually wider in diameter (5-7μm) and slow growing with approximately right angle branching whereas the hyphae of Pythium species are fast growing in general, narrower (4-6μm) wide, with the hyphae more flexuous or meandering on the agar plate. However, the definitive differentiation between them lies in the mode of zoospore differentiation and discharge. In Phytophthora species, the zoospores are differentiated within the sporangium proper and when mature, released in an evanescent vesicle at the sporangial apex, whereas in Pythium species, the protoplast of a sporangium is transferred usually through an exit tube to a thin vesicle outside the sporangium where zoospores are differentiated and released upon the rupture of the vesicle. For many years, they have been classified in the same family and same order: Pythiaceae and Peronosporales4,5 and were commonly referred to as Pythiaceous fungi.
There were attempts to separate them so that Pythium would be placed in family Pythiaceae whereas Phytophthora would be assigned to a new family Phytophthoraceae.6,7 However, this proposal has not received much support from the research community. On the other hand, recent molecular phylogenetic studies suggested that Phytophthora might be more closely allied with downy mildews like Plasmopara Schröter and Peronospora Corda.8,9 Thus, Kirk et al.,10 placed Phytophthora in family Peronosporaceae of order Peronosporales while retaining Pythium in family Pythiaceae of order Pythiales. In view of the fact that insufficient number of Phytophthora and Pythium species and none of the “unculturable” species of Phytophthora have been subjected to molecular analysis it might be premature to separate them from family Pythiaceae that they normally belong to. Many species of these two genera are potentially important plant pathogens11,12 and thus of great economic importance.
The genus Phytophthora (Gr. Phyton, a plant; phthora, destroyer) with 313 described species (www.mycobank.org) was established by de Bary in 187613 with P. infestans as the type species. He recognized that the fungus causing late blight of potato in Europe in the 1840s, previously identified as Botrytis infestans Mont.,14 later Peronospora infestans (Mont.) Casp.,15 had unique conidiophore characteristics, which he considered distinct enough to be assigned a new genus name.
Species of Phytophthora can be readily identified by the aseptate hyphae with slight constriction at the base of its initially right-angled branches; by the ovoid, obpyriform to limoniform sporangia produced in succession on a typically sympodially branched, undifferentiated sporangiophore of indeterminate growth; by the differentiation of mature, laterally biflagellate zoospores within the sporangium proper; by its globose oogonium with single spherical oospore with thin or no periplasm and by antheridial configuration being amphigynous and/or paragynous.16 Most species of Phytophthora are soil-borne plant pathogens causing diseases of both herbaceous and woody mostly dicotyledonous plants, e.g. root rot, basal stem rot, leaf spot or blight as well as fruit rot. They can infect both cultivated and natural plants world-wide causing serious loss in agriculture and upsetting natural forest ecosystem, some of which have attained the status of major epidemics.17–19 Numerous new species were recently isolated from forests, streams and irrigation water but their pathogenicity have not been determined. Although Phytophthora species are well known for causing diseases of terrestrial plants, Govers et al.,20 reported recently the widespread infection of a common seagrass species, Zostera marina (eelgrass) across the northern Atlantic and Mediterranean by P. gemini Man in’t Velf, K. Rosend, Brouwe & de Cock, P. inundata Brasier, Sánch, Hern & Kirk and an unidentified species of Phytophthora threatening the conservation and restoration of vegetated marine coastal systems. The taxonomy of Phytophthora species has been traditionally based on morphological characters, e.g. the papillation, size, length/breadth ratio and caducity of sporangia as well as the ability to produce oospores in single cultures, antheridial configuration and oogonial size21–23 with the six morphological groups of Phytophthora created by Waterhouse22 widely accepted by researchers.
Hyphal swellings are not cut off by septa and when present may be single or clustered, terminal, intercalary or branching to form a network; spherical, oval to irregular, and vary considerably in size. They may be formed on agar plates but more commonly in water. Chlamydospores are sometimes confused with spherical hyphal swellings but they can be distinguished by a secondary wall, the presence of delimiting septum, and usually the internal reorganization of cytoplasm. They are hyaline to yellowish or brownish, thin to thick-walled (mostly 1-2µm), terminal or intercalary.
There are three basic types of sporangia, based on the characteristics of the apex: (a) conspicuously papillate with hemispherical apical thickening over 3.5µm deep (b) inconspicuously papillate (semi-papillate) with shallow apical thickening under 3.5µm deep, and (c) non-papillate, without noticeable apical thickening. A plug may be formed in the sporangiophore, at the base of a papillate or semi-papillate sporangium. Just prior to zoospore release or when the non-papillae sporangia are exposed to air a slight thickening might be detected in non-papillate sporangia which thus appear as “semi-papillate”. The papillate and semi-papillate sporangia may be formed on agar media or host tissues, non-deciduous or deciduous in water with pedicels of variable length, and have narrow exit pores (under 7µm) after the dissolution of the apical thickening. The non-papillate sporangia are usually formed only in water, non-deciduous, have wide exit pores (often over 10µm) and the base of sporangium is not plugged so that the sporangiophore is capable of internal proliferation by producing a new sporangium “nested” within or extended on a new sporangiophore beyond the exit pore of the empty sporangium which often collapses after zoospore dehiscence. The process of internal proliferation can be repeated several times. Laterally biflagellate zoospores are formed within the sporangium and released in an evanescent vesicle, with the dissolution of the apical thickening for papillate and semi-papillate sporangium or inflation of the apex for non-papillate sporangium. The two flagella are inserted in the middle of a longitudinal groove of the ovoid zoospore (10-17x7-12µm), with the anterior end slightly tapering while the posterior end roundish. The zoospore shape varies depending on the angle of observation, appearing as reniform or kidney-shaped only when viewed from the posterior end of the zoospore. The shorter anterior flagellum propels the zoospore with the longer, posterior flagellum trailing behind, serving as the rudder, and it glides smoothly in a spiral fashion while rotating clockwise along its longitudinal axis. In water, after a period of motility which can last up to 24hr or more, the naked zoospore rounds up to form a spherical cyst (7-14µm diam) with a cell wall while the flagella coil up and are discarded. An encysted zoospore may germinate by a germ tube, with or without a germ sporangium or by another zoospore, a process called “repeated emergence”. Occasionally, the sporangium can also germinate directly by one or more than one germ tubes at or near the apex, or close to the sporangiophore. The sporangium has been described as limoniform, obpyriform, ovoid or ellipsoidal, usually terminal, single, basally attached on an undifferentiated, unbranched or sympodially (close or lax) to irregularly branched, narrow sporangiophore (1-3µm) wide. Occasionally, intercalary sporangia may be produced.
Phytophthora species can be either homothallic producing abundant sex organs readily in single cultures or heterothallic, requiring pairing in dual cultures with appropriate A1 or A2 mating types. Those isolates forming oospores with the A2 tester were designated as A1; those forming oospores with A1 tester, A2; those forming oospores with both A1 and A2 testers, A1A2; those that failed to form oospores, A0 (sterile or neuter). The oogonium is mostly terminal, globose to pyriform, hyaline, yellowish to brown, smooth but sometimes uneven or verrucose. The oospore is single, spherical, smooth, hyaline but sometimes yellowish; aplerotic leaving a conspicuous or an inconspicuous space between the relative thick oospore wall (1-3µm or more) and the thin oogonial wall. In the strictest sense, oospores of Phytophthora are rarely plerotic although the term has been used by some reseachers to refer to the condition when the oospore nearly fills the oogonium. There is usually one antheridum per oogonium. The antheridium may be paragynous, applied at the side of the oogonium, often near the oogonial stalk or amphigynous, encircling the oogonial stalk. The amphigynous antheridium may be unicellular or bicellular. The amphigyny is the result of penetration of the antheridial initial by the oogonial initial during the early stage of sexual reproduction.24 The amphigynous antheridium is a unique feature of Phytophthora, shared only with the monotypic genus Trachysphaera Tabor & Bunting.25 However, with the advent of molecular biology and automation, the concept of taxonomy has quickly changed so that Gallegly & Hong26 used a combination of morphology and DNA fingerprints in their new key to Phytophthora whereas Ristaino27 introduced a lucid key to the common Phytophthora species based on a series of interactive computer matrixes of morphological and molecular data. Attempts were made to develop a cyberinfrastructure to facilitate the identification of Phytophthora species based solely on molecular data. Thus, Park et al.,28 used the Phytophthora sequence database to develop a website: (www.phytophthoradb.org), whereas Grünwald et al.,29 introduced another website: (www.Phytophthora-ID.org). Furthermore, there was a movement to shift from the six artificial morphological groups of Waterhouse22 to the establishment of phylogenetic clades of Phytophthora based on the molecular data to reflect a natural classification of special note is the establishment of eight clades established by Kroon et al.,30 to be followed by ten clades by Blair et al.,31 and the existence of 10 clades among Phytophthora species is now well recognized.32,33 The morphological characteristic of the genus Phytophthora are illustrated by Figure 1. Terms used were defined by Blackwell.34
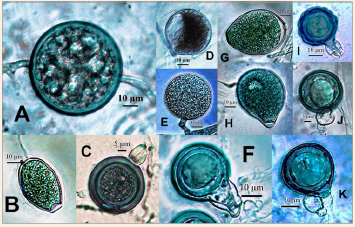
Figure 1 Phytophthora. A. Thick-walled chlamydospore. B. Semi-papillate sporangium. C, F. Amphigynous antheridia with smooth oogonia. D. Hyphal swelling. E. Thin-walled chlamydospore. G. Non-papillate sporangium. H. Papillate sporangium. I. Bicellular amphigynous antheridium with verrucose oogonium. J, K. Paragynous antheridia with smooth oogonia.
The genus Pythium Pringsh., with 355 described species (www.mycobank.org), has been notoriously difficult for its taxonomy. Historically, there has also been great confusion regarding the validity of Pythium as a distinct genus. The genus was created by Pringsheim in 185835 and placed in the family Saprolegniaceae. However, Pythium Pringsh. was antedated by both Pythium Nees and Artotrogus Montagne.36 Subsequently, proposals were made to conserve the genus Pythium Pringsh. against Pythium Nees37 and Astotrogus Montagne38 so that Pythium Pringsh became the de facto official genus name. However, there were attempts to split the genus Pythium into two genera to differentiate species with spherical sporangia from those with filamentous sporangia. Schröter39 erected the family Pythiaceae in which he described Pythium having globose sporangia and Nematosporangium with filamentous sporangia. On the other hand, Sparrow40 proposed species of Pythium with globose sporangia to be placed in a new genus, Sphaerosporangium.
Others tried to create various infrageneric taxa within Pythium but all these proposals have been rejected by Waterhouse,41 Plaats-Niterink,12 and Dick.42 Nevertheless, recent phylogenetic studies based on molecular data have provided new evidence and rekindled interest to split the genus Pythium. Thus, Ko et al.,43 erected a new genus Aquaperonospora Ko for species producing rigid, erect and branched sporangiophores forming sporangia synchronously on branchlet tips. Bala et al.,44 proposed a new genus Phytopythium Abad for those species with globose to ovoid, often papillate and internally proliferating sporangia. Uzuhashi et al.,45 restricted the genus Pythium to those species with inflated or non-inflated filamentous sporangia while creating four new genera to accommodate species with non-filamentous sporangia: Ovatisporangium Uzushashi, Tojo & Kakish for species with mainly ovoid to pyriform, sometimes irregular shaped-sporangia, Elongiosporangium Uzushashi, Tojo & Kakish for species with clavate to elongate sporangia, Globisporangium Uzuhashi, Tojo & Hahish. for species with globose, sometimes proliferating sporangia and Pilasporangium Uzushashi, Tojo & Kakish. for species with globose, non-proliferating sporangia. Based on molecular phylogenetic analysis, de Cock et al.,46 distinguished species of Pythium into 14 clades: A-K. All species in Clade K, including Py. boreale Duan, Py. chamaehyphon Sideris, Py. cucurbitacearum Takimoto, Py. helicoides Drechsler, Py. indigofereae Butler, Py. oedochilum Drechsler and Py. vexans de Bary were re-classified as species of Phytopythium44 which is morphologically intermediate between Phytophthora and Pythium, with priority over and treated as synonymous with Ovatisporangium.46 In distribution, the species of Pythium are cosmopolitan, widely distributed throughout the world ranging from tropical to temperate12 and even arctic47 and antarctic regions.48
Economically, they are especially important as pathogens of monocotylodonous plants, causing serious damage to cereal crops and turf grasses, leading primarily to soft rot of fruit, rot of roots and stems, and pre- and post-emergence of seeds and seedlings by infecting mainly juvenile or succulent tissues.49 Whereas Pythium diseases are common in tropical to temperate regions, cereal seedlings under snow could also be killed by Pythium spp.50,51 and an unidentified species of Pythium isolated from a colony of diseased leafy liverworts from Signy Island of Antarctic proved to be a potential plant pathogen to local vascular plants, based on artificial inoculation.52 Other species caused diseases in fish,53 marine red algae54,55 and mammals including humans, causing pythiosis.56,57 On the other hand, some species of Pythium can produce valuable compounds for pharmaceutical and food industry, like polyunsaturated fatty acids, arachidonic acid and cicospentaenoic acid,58 synthesize biotin, folic acid, pantothenic acid, riboflavin, thiamine and vitamin C, etc. and are capable of transforming sterols into cholesterol, ergosterol and sitosterol whereas the copious production of hydrolytic enzymes like pectinase, amylase and cellulase. might find uses in industry, for instance, in the breakdown of plant wastes for fermentation to produce biofuel.59 There are some mycoparasitic species of Pythium, capable of controlling plant diseases due to other fungi.60,61 In fact, a pesticide has been developed from Pythium oligandrum and marketed for seed dressing, overhead spraying and soil drenches. In addition, other species of Pythium are entomopathogenic and can be used as biological control of mosquitoes and other harmful insects.62,63
The identification and taxonomy of Pythium species has always been difficult due to the paucity of stable morphological characters despite the extensive studies in the early days by Mathews,64 Sideris65 and Middleton.66 Waterhouse41 and Dick42 provided useful identification keys to the genus Pythium. Furthermore, Waterhouse67 compiled the original descriptions of all Pythium species up to that time, but it was Plaats-Niterink12 who wrote the most complete monograph on Pythium with a user-friendly identification key to 85 species which was well received by researchers. However, with the shape of sporangia and ornamentation of the oogonia serving as the primary diagnostic characters supplemented with other features including the size of oogonia and oospores, the extent of oogonial space occupied by the oospores, the origin, shape, number and method of attachment of antheridia to oogonia and the presence of hyphal swellings, the identification of Pythium species still proves to be a challenging task due to the infraspecific variability and interspecific overlapping of these morphological data and the numerous new species reported since 1981. There is an urgent need for an updated identification key to include all species of Pythium described. Bouket et al.,68 introduced a web-based identification program for 90 Pythium species based on morphological features. As in Phytophthora, molecular tools have become increasingly important in species differentiation of Pythium.69 However, the large amount of molecular data obtained by many researchers have not yet been organized to provide a database website for species identification as in the case of Phytophthora. The phylogeny of Pythium based primarily on the sequencing of ITS region of the nuclear ribosomal DNA of 116 species and varieties revealed the existence of 11 major clades, designated as Clades A-K,70 approaching a natural classification of the genus Pythium.
The sporangia of Pythium are usually produced in water and non-caducous. Some species have papillate sporangia but they lack the apical thickening characteristic of papillate sporangia in Phytophthora. The protoplasm of the sporangium is discharged through an exit tube to form a vesicle at the end where zoospores are differentiated and released upon the rupture of the vesicle. Morphologically similar to Phytophthora, zoospores of Pythium are laterally biflagellate and encyst after a period of motility to germinate into 1 or more than 1 germ tubes that will develop into mycelia. The sporangia can be filamentous, indistinguished from the rest of the mycelium, or slightly inflated to form dendroid structures or greatly inflated to form lobulate or toruloid complexes. In contrast, the sporangia can be globular, subglobular to ovoid, with or without internal proliferation; some may have apical papilla but lacks the apical thickening characteristic of those species of Phytophthora producing papillate or semi-papillate sporangia.
The sexual reproduction is by means of oogonia and antheridia. The oogonia can be globular to limoniform, terminal or intercalary on undifferentiated hyphae with the oogonial wall smooth or ornamented with projections of various length and evenly or unevenly distributed. The unicellular antheridia can be sessile or intercalary on the hyphae or terminal on a special stalk; monoclinous if they originate from the oogonial stalk, diclinous if they originate from a different hypha not closely connected with the one subtending the oogonium or hyopgynous if the proximal portion of the oognial stalk becomes an antheridium. The oogonium usually contains one or occasionally more than one spherical, smooth oospores which can be aplerotc or plerotic with the entire oogonial space fully occupied by the oospore. The oospore wall can be thin to thick. All species of Pythium with the exception of about seven species (Py. catenulatum Mathews, Py. flevoense Van der Plaats-Niterink, Py. heterothallicum Campbell & Hendrix, Py. intermedium de Bary, Py. macrosporum Vaartaja & Van der Plaats-Niterink, Py. splendens Braun and Py. sylvaticum Campbell & Hendrix) are homothallic, producing sex organs in single cultures.
Hyphal swellings can be present, often indistinguishable from young spherical oogonia or sporangia and are capable of germination to form a new mycelium; terminal, intercalary or catenulate, non-deciduous but can be deciduous in Py. intermedium. Thick-walled chlamydospores are rare, occurring only in Py. dimorphum Hendrix & Campbell and Py. tracheiphilum Matta. Appressoria are slight swellings formed by hyphae, especially when in contact with the walls of the Petri dishes, with various shapes and sizes, ranging from sickle-, club- or sausage-shaped to subglobose, single, terminal or intercalary forming chains or clusters.
The morphological characteristics of Pythium are summarized in Figure 2 & 3.
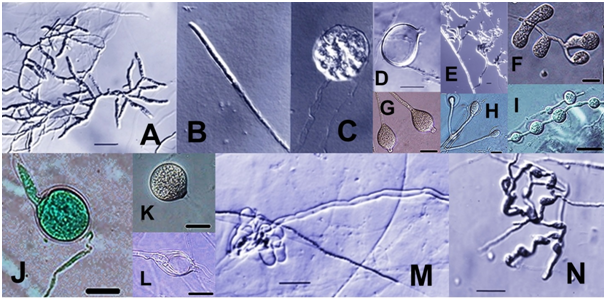
Figure 2 Pythium. A-C. Filamentous sporangia with single sporangium developed from hyphal tip (B) and zoospore formation in a vesicle at the end of an empty sporangium (C). D. Empty bursiform sporangium with short exit tube. F. Lobulate sporangium. G. Papillate globular sporangium. J. Globular sporangium with medium exit tube. H, L. Internally proliferating sporangia. M. Empty lobulated sporangium with long exit tube. E, N. Appressroria. I, K. Interclalary and terminal hyphal swellings. (All scale bars: 20 µm; B,C at the same magnification as D).
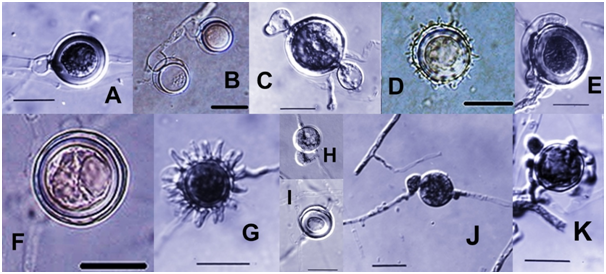
Figure 3 Pythium. A. Plerotic oospore with hypogynous antheridium. B. Aplerotic oospores with intercalary, basal antheridia. C. Oogonium with two crook-neck antheridia. D, G. Ornamented oogonia with conical projections in D and digital projections in G. E. Elongated, curved antheridum attached laterally on the oogonial surface. F, I. Smooth oogonium with aplerotic oospores. H. Oogonium with monoclinous antheridium. J. Oogonium with diclinous antheridium. K. Oogonium surrounded by several antheridia. (All scale bars at 20 µm; H at the samemagnification as I).
Although morphological characteristics will still serve as the basis for identification of Phytophthora and Pythium species12,27,29 it is clear that further studies using various molecular biology techniques are urgently needed to study the phylogeny of as many isolates as possible of all known species of these two economically highly important genera in order to develop a natural classification and rapid tools for identification of the unknown isolates especially when some key morphological characters are lacking. In Phytophthora, useful websites have been developed28,29 to allow species identification by BLAST against the collections in GenBank based primarily on similarity coefficient of the sequential analysis of the rDNA, especially the ITS region. However, the sequences housed in GenBank have not been carefully curated and mistakes could be made but this approach still seems to be the most promising as long as the isolates deposited could be checked for accuracy in identification and other molecular markers used more often by other researchers. In the future, species identification by automation should be the ultimate goal. Some species like P. cyperi and P. polygoni have been considered “unculturable” and attempts should be conducted to develop a medium that can support their growth for studies. Meanwhile, at least they should be compared with other culturable species based on the nucleotide sequence and other molecular markers to determine their relationships. Tyler (personal communication) just completed an analysis of the nucleotide sequence of P. polygoni and found it to form a previously undescribed subgenera clade of Phytophthora. It is of interest to note recent isolations of Phytophthora-like organisms from natural and semi-natural ecosystems in Europe, Chile and Vietnam and based on morphological and phylogenetic studies they were treated a new sister genus of Phytophthora: Nothophyhtophthora gen. nov. Jung et al.71 As for Pythium, no data base has been developed to date to allow species identification based on molecular studies. It would be extremely useful if a contemporary, rigorous website can be constructed in the near future to allow more rapid identification of Pythium species based on molecular data because compared with Phytophthora species, the morphological characters of Pythium are even more complicated and variable.
None.
The author declares no conflict of interest.

©2018 Ho. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.