Journal of
eISSN: 2469 - 2786


Research Article Volume 9 Issue 4
Department of Animal Preventive Medicine, University of Chile, Chile
Correspondence: Navarro C, Department of Animal Preventive Medicine, FAVET, University of Chile, Chile
Received: October 05, 2021 | Published: October 28, 2021
Citation: Vera C, Jara MA, Navarro C. The Fsp region from fusion protein gene of canine distemper virus: high variability? J Bacteriol Mycol Open Access. 2021;9(4):153-161. DOI: 10.15406/jbmoa.2021.09.00315
The Canine Distemper is one of the main infectious diseases in domestic dogs. The introduction of live attenuated-viral vaccines has helped to maintain the disease under control. However, in the past few decades it has been observed worldwide a rising number of cases even in vaccinated animals. The canine distemper virus lineages circulating in the world have been described based on the hemagglutinin analyses, due to its high degree of genetic variability. Recently, new studies have reported greater variations in the amino acidic sequence of a region in the fusion protein. In order to determine the variability of the field strains in comparison with the vaccines and strains from other lines, in this dissertation the genomic variability of the Fsp region from the canine distemper virus fusion protein gene is analyzed. With this purpose, a chain-reaction of the polymerase with reverse transcription, capable of amplifying this variable region, was implemented and identified through its nucleotide sequence. These sequences were compared with vaccine strains and with field strains of known lineages. Additionally, a phylogenetic tree was built for this variable region. The results of the nucleotides comparison show that the field strains have more homology to the vaccine strain Onderstepoort and according to phylogeny, it would belong to the America 1 lineage.
Keywords: canine distemper virus, canine distemper, fusion protein, F protein, Fsp, RT-PCR
The Canine Distemper (CD) is a contagious, lethal, globally distributed multisystem disease caused by the Canine Distemper Virus (CDV). This virus affects mammals of the carnivorous order, members of the families Canidae, Felidae, Mustelidae, Ailuridae, Mephitidae, Procyonidae, Ursidae, Viverridae, Hyaenidae and Phocidae,1–4 being the domestic dogs, most affected by this pathology. In addition, the disease has been observed in collared peccary (Tayassu tajacu) in Arizona,5 and in some primates such as the Japanese macaque (Macaca fuscata) in Japan and Rhesus monkeys (Macaca mulatta) in China.6,7 DC is a disease that affects dogs of all ages, with puppies from 3 to 6 months of age being the most susceptible because they have lost maternal antibodies and their immune system is still immature to respond to infection.8 The main routes of entry of the virus are ocular, respiratory and oral, through aerosols and fomites, reaching mucosal surfaces where the first interaction with the host’s immune system is established by the early infection of local lymphocytes and mononuclear cells.3,9 The first viremia occurs between days 4-6 post-infection (PI) and produces infection of the lymphatic tissues with the onset of fever and the onset of lymphopenia. The second viremia is accompanied by pyrexia and determines the infection of the epithelial cells of all tissues of the body. This viremia is accompanied by the appearance of rashes and hyperkeratosis and determines the beginning of the symptomatic phase.4,8,10 The clinical signs associated with the disease depend on the virulence of the viral strain, the age, immune and health status of the animal. In susceptible species, the respiratory, gastrointestinal and central nervous system (CNS) systems are the most compromised. The highest percentage of infections in domestic dogs is subclinical, presenting the disease mildly and with symptoms such as languor, anorexia, one fever and infection of the upper respiratory tract. On the other hand, the acute form of the disease presents a high mortality, manifesting signs associated with the respiratory and gastrointestinal system, including conjunctivitis, oculo-nasal discharges, pneumonia, vomiting, diarrhea (often hemorrhagic), cachexia and severe dehydration. Depending on the viral strain, acute encephalomyelitis may occur in association or immediately after systemic manifestation. Among the numerous neurological manifestations associated with infection (cervical stiffness, seizures, vestibular and cerebral symptoms, and sensory ataxia), myoclonus is the only neurological symptom suggestive of DC.1,4,8,11,12 Severe immunosuppression associated with CDV enhances the susceptibility to secondary infections, contributing to the high death rates of the disease.9
Currently, vaccination is the main strategy to control the disease. Vaccines with live attenuated virus (LAV) stimulate the humoral and cellular immune response and induce immunological memory. The development and use of these vaccines have contributed to a drastic reduction in the incidence of CD in domestic dogs. In spite of this, outbreaks of the disease have been observed recently in populations of immunized dogs belonging to different geographical regions. Although the causes have not been determined, these outbreaks could be explained either by the reversion of the attenuated strains to the virulence, by the emergence of new strains sufficiently variable to evade the immune response generated by the vaccines, by failures in the administration of vaccines, or because of the sanitary and immunological status of the animal.2,8,13–17 At present, the diagnosis of CD is based on clinical suspicion supported by the manifestation of clinical signs and in the risk, antecedents predisposing to the disease. However, the lack of specificity in the symptoms associated with the infection can lead to confusion with other pathologies in the final diagnosis, which is why two developed various complementary diagnostic methodologies, including serological techniques, such as immunohistochemistry and ELISA (Enzyme-Linked Immune Sorbent Assay); and molecular techniques, such as the RT-PCR assay (Polymerase Chain Reaction after reverse transcription).4,8,10 Although the measurement of serum antibodies (Ac) IgM (against the viral core proteins N and P) and IgG (against the antigens of the H and F proteins) can help in the diagnosis of the virus, the problems of the serological tests they reside in not being able to differentiate between maternal, vaccinal, or infection-infected.18 Thus, the detection of Ac is not sufficient for the definitive diagnosis, given that infected dogs not vaccinated with acute presentation can die without the appearance of neutralizing Ac, while those infected in a subacute or chronic way can have Ac levels comparable with the vaccinated dogs. This is why molecular methodologies based on RT-PCR have been developed by means of the amplification of nucleic acid fragments, to make a precise ante- mortem diagnosis in presumptive cases of CDV infection and can be applied to the study. of genomic variations of the virus through the molecular characterization of relevant sequences of the genome.8,18,19
CDV belongs the Mononegavirales order, Paramyxoviridae family and Morbillivirus genus. It is closely related to the human measles virus, the morbilliviruses of cetaceans, the rinderpest virus and small ruminants. It was described by Jenner in 1809, but its viral etiology was demonstrated by Henri Carré in 1905.4,8,14,20 It is a enveloped virus of size between 150 to 300 nm in diameter and its genome is approximately 15.7 kilobases (kb) in length and is composed of unsegmented, single-stranded ribonucleic acid (RNA), sense of negative coding and encodes 6 viral proteins: the nucleocapsid protein (N), the phosphoprotein (P), the polymerase protein major (L), the matrix protein (M), the hemagglutinin (H) and the fusion protein (F).21–24 The lipid envelope contains the two surface glycoproteins (F and H) that mediate the recognition, entry and exit from the target cell, both being the objective of the neutralizing antibodies generated by the host’s immune system. Protein N encapsulates viral RNA and together with proteins P and L constitute the ribonucleoprotein complex (RNP), which initiates the replication and transcription of the viral genome. The M protein connects the surface glycoproteins and the nucleocapsid during viral maturation.4,11,24
Protein F is a transmembrane glycoprotein that participates in the fusion of the viral envelope with the plasma membrane of the host cell.3 The F protein is encoded by the F gene in the form of an inactive precursor called pre-F0, which, by recognition of the signal peptide (Fsp), is processed to form the F1 and F2 subunits that establish a heterodimer to constitute the active form of protein F (Figure 1). Fsp is not found in mature viral particles because the maturation of the F protein requires its proteolytic processing. In spite of the above, it would be indirectly involved in the activity of the F protein, limiting its expression at the intracellular level and on the surface, with the consequent reduction of cell-cell fusion levels. This region would also contribute to virulence and even to viral pathogenesis.15,16,21,23,24 The F gene is the second most variable segment within the CDV genome. At the level of its amino acid sequence, F protein varies around 4% between field isolates, F1 and F2 subunits being more conserved, while the Fsp region shows variations of up to 27%.24,25 Regarding the differences between nucleotides, in a study by Sarute26 a range of 65-69 variable nucleotide sites was observed when comparing field sequences from Uruguay and Argentina with the vaccinal strain Onderstepoort.26 Therefore, the variability values of the Fsp region are even higher than those described for protein H (around 10%), which is why this region has been used in CDV characterization and evolution studies.23,24,25,27 For the Fsp region phylogeny studies have also been carried out, finding agreement with the existing lineages for protein H, except for the African lineage because it has not yet been characterized by this region. CDV lineages The CDV lineages circulating in the world have been described based on the analysis of hemagglutinin because it has a high degree of genetic heterogeneity, allowing up to date, to classify the majority of field strains in eight lineages: Africa, America- 1 (vaccine strains), America-2, Asia-1, Asia-2, Europe-1, Europe-2 (Europe wild-life) and Europe-3 (Arctic-like).28 However, new studies have detected greater variations in the amino acid sequence of a region of the fusion protein (F).17,25,27
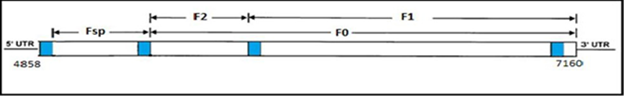
Figure 1 Scheme of the F gene (nucleotides 4858-7160). Regions that code for the subunits of the F protein.23
Phylogenetic studies in Uruguay: both H gene and the Fsp region, have described two field lineages currently co-circulating in South America: the so-called Europe1/South America1, which extends into Europe, Argentina, Brazil and Uruguay; and South America 2, which is exclusive to Argentina.2,27 Added to this, other studies conducted in 2014 reveal the existence of two new variants of the virus: one found in samples from Ecuador, classified by17 in the South American lineage 3 by phylogenetic analysis of the Fsp region; and another belonging to Colombia, also classified in the South American lineage 3 by10 through the analysis of the H gene. As both variants were classified by different regions, it cannot be established whether there is a relation between them.10,17 This has led to the description of at least 14 CDV genotypes. Independent of the above, in our country it determined through the study of a segment of the H gene that there would be at least two lineages circulating: America 1 and Europe 1.28 The objective of this work is to perform a comparative analysis and phylogeny of the nucleotide sequences of some field strains present in the metropolitan region and known sequences of the variable region of the F protein gene, to preliminarily determine the genomic variation of the Fsp region. Of the CDV F protein gene.
The present work was carried out in the Virology and Microbiology laboratories of the Department of Animal Preventive Medicine of the Faculty of Veterinary and Animal Sciences of the University of Chile, with financing from FIV PROJECT 121014019102010.
Controls
As a negative control, blood was used with anticoagulant of dogs that did not present clinical signs of the disease, without vaccinations and without risk antecedents. For the positive controls, commercially available vaccines were used with VVA from CDV, "Nobivac® Puppy DP" (Onderstepoort strain), "Canigen MHA2PPi / L" (Lederle strain) and "Vanguard® Plus 5/ L" (Snyder -Hill strain). For the control of the reagents, nuclease-free water was used.
Samples
We used 22 isolates of CDV genomic RNA previously extracted from peripheral blood, collected from the Veterinary Neurological Institute in tubes with EDTA (2mL), from domestic dogs that showed clinical signs of the disease, some with positive serology to CDV and positive to the detection of the CDV H and N gene. These samples were stored more than a year ago in Virology and Microbiology laboratories at -20°C.
Obtaining viral RNA (vaccines)
Viral genome extraction from commercial vaccines was performed using the TRIzol® LS extraction kit. For this, each lyophilized vaccine was reconstituted with 750μL of reagent, allowing it to incubate for 5 minutes at room temperature (RT). Then, 200μL of chloroform was added to each tube, mixed vigorously for 15 seconds and incubated at RT for 5 minutes. Subsequently, the tubes were centrifuged at 7000xg for 15 minutes and the aqueous phase was transferred to clean tubes. For RNA precipitation, 0.5mL of isopropanol was added to each tube, left at RT for 10 minutes, centrifuged at 7000xg for 10 minutes, the supernatant was removed, washed 3 times with ethanol (1mL of 75% ethanol), vortexed for 15 seconds and centrifuged at 2000xg for 5 minutes at RT. Once the centrifugation was finished, the supernatant was removed, the RNA precipitate could dry in vacuo for 5 minutes and resuspended in 100μL of nuclease-free water. Finally, the RNA was incubated at 60°C for 10 minutes and then stored at -20°C until further use.
Primers design
The primers for the reaction were designed by using the free access online program OligoPerfectTM Designer from Life TechnologiesTM, and they were synthesized to Bioscan®. For the design, the nucleotide sequence of the region comprising nucleotides 4858-7160, corresponding to the complete F protein gene of the Onderstepoort vaccine strain (AF378705) available in GenBank®.24 Then the desired parameters for these starters were selected: particle size between 18-30 base pairs (bp), melting temperature 55-60°C% GC between 40-60%.29 The size of the amplicon was between 490-510 bp to encompass the complete Fsp region of 405 bp of the F gene (4935-5340 nt) (Figure 2). The program it gave us the following pair of primers, P1: 5'-GACAGGAACCCCCACAAAC-3' (4905-4923nt) and P2: 5'-GATCTTGTAATGGACACTATCAGTCC-3 '(5381-5406 nt), those that allowed to amplify a fragment of around 500bp. RT-PCR: To perform the RT-PCR, the Apollo thermocycler (CLP, USA) of 96 wells of 0.2mL was used and a protocol with appropriate temperatures, times and cycles for each stage.
Reaction mixture
The RT-PCR reaction was performed using the "MyTaq™ one step RT- PCR" kit (Bioline®) following the manufacturer's instructions. To each tube were added 16μL of "2X Reaction Mix", 0.5μL of RNase-inhibiting "RiboSafe", 0.5μL of reverse transcriptase, 5μL of each primer and 5μL of RNA annealing obtaining a final volume of 32μL. In the thermocycler the following protocol of cycles and temperatures for the reaction was used: 1 cycle at 4°C for 30 minutes and at 95°C for 1minute to perform the reverse transcription and activate the polymerase respectively; 30 cycles to perform the amplification at 95°C for 10 seconds (denaturation), 55°C for 10 seconds (alignment), 72°C for 30 seconds (elongation) and 1 cycle at 72°C for 5min (final extension). Once the thermocycler process was finished, the PCR tubes were stored at 4°C.
Visualization of the amplified products
To visualize the RT-PCR products, 2% agarose gel electrophoresis was performed in Tris HCl buffer (100mM Tris-HC1, 10mM EDTA). The product of each RT-PCR was mixed with a commercial loading product (Fermentas®) and loaded into the wells of the gel, also a molecular size "O`Range RulerTM" (Fermentas®) marker (Fermentas®) was added to a well. Contains fragments of DNA between 50 and 1000 bp, which helped determine the size of the amplified fragment. The electrophoresis was performed at 90V for 45 minutes. After, the gel was incubated in ethidium bromide (0.5μg/mL) for 60 minutes, then left for 10 minutes in water to remove excess reagent. Once this time was finished, it was visualized in a transilluminator of ultraviolet light and was photographed.
Sequencing of amplified fragments
Sequencing Center of the company Genytec Ltda. 4 of the samples that were positive to the method were sent in triplicate. Within the selected samples, 2 correspond to RNA isolates that were previously used to perform the phylogenetic analysis of haemagglutinin.28
Determination of nucleotide identity
The obtained sequences were aligned using the open access oanline program Clustal Ω, obtaining a consensus sequence for each sample used. Then, these consensus sequences were entered to the open access online program BLAST to identify the origin of DNA fragments amplified in RT-PCR.
Comparison of nucleotide sequences
The consensus sequences of the 4 samples were aligned using the Clustal Ω program and compared with each other to determine the percentage of existing genomic variation. Then they were compared with the sequences of the vaccinal Onderstepoort and Snyder Hill strains, and later with some known sequences of isolates belonging to the different lineages.
Phylogenetic analysis
The phylogenetic analysis of the obtained sequences was carried out through the MEGA 6.06 program of bioinformatic analysis. For this, the 4 amplified sequences and some reference sequences of the previously known lineages were entered the program (Table 1). Through the methods of "neighbor-joining" and "maximum likelihood", two phylogenetic trees were constructed for each method using different evolutionary models (four in total). After analyzing the data, the tree that was developed was chosen 10 through the method of "neighbor-joining" with the evolutionary model of "p-distance" to calculate the existing genetic distances. The robustness of the tree was determined by the analysis of "bootstrap" using a thousand replicas.30
Strain |
Origen |
Access number |
Lineage |
Onderstepoort |
Vacuna/EEUU |
AF378705 |
América 1 |
Snyder Hill |
Vacuna/Canadá |
GU138403 |
América 1 |
A75/17 |
Suiza |
AF164967 |
América 2 |
164071 |
EEUU |
EU716337 |
América 2 |
01-2689 |
EEUU |
AY649446 |
América 2 |
5804P |
EEUU |
AY386316 |
Europa 1 |
Arg23 |
Argentina |
KC257465 |
Europa 1 |
UY102 |
Uruguay |
KC331150 |
Europa 1 |
19876 |
EEUU |
AY964110 |
Europa 2 |
18133 |
EEUU |
AY964108 |
Europa 3 |
HL |
China |
EF596901 |
Europa 3 |
21261 |
EEUU |
AY964112 |
Europa 3 |
W729B |
Japón |
AB607904 |
Asia 1 |
GS0812-4 |
China |
HQ850148 |
Asia 1 |
HeB(07)1 |
China |
EU327874 |
Asia 1 |
Th12 |
Tailandia |
AB509344 |
Asia 1 |
TW-KM2 |
Taiwan |
EU192008 |
Asia1 |
007Lm |
Japón |
AB474397 |
Asia 2 |
007Lm-1vp |
Japón |
AB462810 |
Asia 2 |
M25CR |
Japón |
AB475097 |
Asia 2 |
011C |
Japón |
AB476401 |
Asia 2 |
50Con |
Japón |
AB476402 |
Asia 2 |
Arg24 |
Argentina |
KC257466 |
Sudamerica |
Arg25 |
Argentina |
KC257467 |
Sudamerica |
Table 1 Nucleotide sequences used in comparative and phylogenetic analyzes
Analysis of results
Those samples that after the RT-PCR originated a fragment of DNA of around 500 bp were considered positive and their nucleotide identity was confirmed with CDV when entering them into the BLAST program. The results of the comparison of nucleotides were analyzed according to the data of the patients (sex, age, race, vaccination status, clinical signs) to determine if there were variations that could be associated with these parameters. In addition, through eleven Comparison of field strains with commercial vaccines was evaluated if existing differences could explain the ineffectiveness of the latter in the immunity of the dogs. The construction of the phylogenetic tree allowed us to know the lineage of the 4 sequenced samples, according to the Fsp region of the F gene. At the same time, the phylogeny results obtained in the phylogenetic analysis of the protein H gene were compared,28 with those achieved for the Fsp region.
Biosafety measures
To carry out the laboratory work, safety measures were implemented according to the biosafety levels established for the Microbiology and Animal Virology laboratories, consisting of the use of clean material, use of white apron and gloves. In addition, for the visualization of the amplified products, due to the handling of ethidium bromide and the transilluminator of UV light, the use of an acrylic plate and glasses with UV filter was required, the latter to protect the vision of the observer. On the other hand, both the gel that was immersed in ethidium bromide and the gloves used were incinerated since this compound has mutagenic properties.
Once the designed primers were synthesized and the RT-PCR protocol was established in 30 cycles and alignment temperature at 55°C. To visualize the results, the samples were separated into 2gels. In the first, the negative controls were loaded; the positive controls corresponding to the commercial vaccines of the Onderstepoort, Lederle and Snyder Hill strains that generated medium intensity bands of around 500 bp; and 12 RNA isolates, of which one originated visible band (Figure 3).
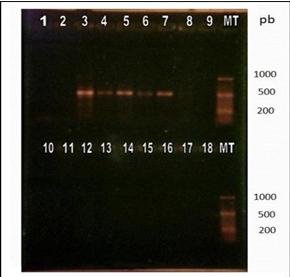
Figure 3 Visualization of products amplified by RT-PCR by electrophoresis in 2% agarose gel with subsequent incubation in ethidium bromide.
It is important to note that the canine that tested positive for this method had had two blood samples taken, both positive to CDV by RT- 12 PCR of the N. gene. The one that produced the band was processed on the same day of the blood extraction; the one that did not produce a band was processed three days later. In the second gel, the negative controls were loaded, 10 RNA isolates positive to the RT-PCR methods for H and N genes that originated bands of around 500 bp corresponding to the expected DNA fragment, and the control of reagents (Figure 4).
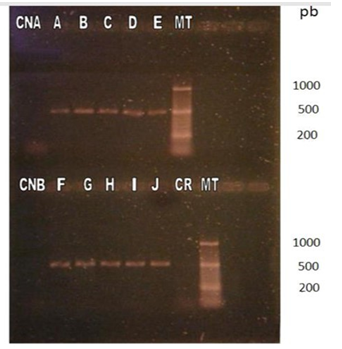
Figure 4 Visualization of products amplified by RT-PCR by electrophoresis in 2% agarose gel with subsequent incubation in ethidium bromide.
Sequencing
The nucleotide alignment for each sample (Annex 1) allowed to obtain four consensus sequences: CDV/CVY1 (canine 7), CDV/CVY2 (canine A), CDV/CVY3 (canine and CDV/CVY4 (canine H) (Table 2). Subsequently, to identify the origin of the amplified DNA fragments in the RT-PCR, the consensus sequences were entered the BLAST program and it was verified that the four sequences correspond to CDV (Annex), corroborated by the high percentages of nucleotide identity obtained with respect to the official sequences stored in the GenBank®.
|
> CDV/CVY1 |
|
ATGCACAAGGGAATCCCCAAAAGCTCCAAAACCCAAACATACACCCAAC |
|
> CDV/CVY2 |
|
ATGCACAAGGGAATCCCCAGAAGCTCCAGAACCCAGACACATGACCAACAA |
|
> CDV/CVY3 |
|
ATGCACAAGGGAATCCCCAAAAGCTCCAAAACCCAAACACTTACCCAACAA |
|
> CDV/CVY4 |
|
ATGGATAAGGGAATCCCCAAAAGCTCCAAAACCCAAAGAGAGACCCAACAAGACCGCCCCCGATAACCCAGCACCGAACCCGAAGAGACCAGGACCTCCCGAGCACGAGATAGCATAACATCAGCTCAGCGATCCACGCACTATGATCCTCGAACATCGGAGATACCCGTCTCCTACACCATGGACAGGATCAGGTCCCGCAAGCAAACTAGCGATAGATTGAAGAACATCCCAGTTCACGGAAACCACGAGGCTATTATCCAGGAGAGACGATAGAGTGTCTCAAAAGGAGGAGATCCGATATCGAAAGGCGGCAACCCAATGCAATCAACTCAGGCTCTCAGTGCACCTGGTTAGTCCTGTGGTGCCTCGGAATAGCCAGTCTCTTTCTGTGTTCCAAGGCT |
Table 2 Consensus sequences for the 4 amplified samples aligned by means of the Clustal Ω program. These sequences were edited from the results from the amplified fragment of around 500 bp, conserving only the 405 bp corresponding to the Fsp region of the F gene of the CDV
Comparative sequence analysis: The consensus sequences indicated in Table 4 were entered the Clustal Ω program to perform the comparative analyzes.
|
|
NIP |
|||
|
Strain |
CDV/CVY1 |
CDV/CVY2 |
CDV/CVY3 |
CDV/CVY4 |
|
CDV/CVY1 |
100 |
83 |
86 |
84 |
|
CDV/CVY2 |
83 |
100 |
88 |
87 |
|
CDV/CVY3 |
86 |
88 |
100 |
89 |
|
CDV/CVY4 |
84 |
87 |
89 |
100 |
Table 3 Percentage of identity of the field sequences
|
Strain |
NIP |
|
|
Onderstepoort |
Snyder hill |
|
|
CDV/CVY1 |
90 |
85 |
|
CDV/CVY2 |
93 |
88 |
|
CDV/CVY3 |
94 |
89 |
|
CDV/CVY4 |
95 |
90 |
Table 4 NIP of field sequences respect onderstepoort and snyder hill strains (commercial vaccines)
|
Lineage/strain |
01 |
02 |
03 |
04 |
05 |
06 |
07 |
08 |
09 |
10 |
11 |
12 |
|
01: Asia 2 |
100 |
87 |
86 |
89 |
88 |
88 |
87 |
85 |
81 |
80 |
79 |
77 |
|
02: Europa 3 |
87 |
100 |
84 |
86 |
86 |
86 |
87 |
84 |
80 |
79 |
79 |
75 |
|
03: Asia 1 |
86 |
84 |
100 |
88 |
88 |
88 |
87 |
82 |
79 |
77 |
77 |
75 |
|
04: America 2 |
89 |
86 |
88 |
100 |
90 |
90 |
89 |
86 |
81 |
80 |
81 |
78 |
|
05: Europa 1 |
88 |
86 |
88 |
90 |
100 |
90 |
89 |
84 |
80 |
78 |
79 |
76 |
|
06: Sudamerica |
88 |
86 |
88 |
90 |
90 |
100 |
90 |
84 |
80 |
79 |
79 |
78 |
|
07: Europa 2 |
87 |
87 |
87 |
89 |
89 |
90 |
100 |
84 |
80 |
79 |
80 |
77 |
|
08: America 1 |
85 |
84 |
82 |
86 |
84 |
84 |
84 |
100 |
95 |
93 |
94 |
90 |
|
09: CVY4 |
81 |
80 |
79 |
81 |
80 |
80 |
80 |
95 |
100 |
88 |
89 |
86 |
|
10: CVY2 |
80 |
79 |
77 |
80 |
78 |
79 |
79 |
93 |
88 |
100 |
87 |
83 |
|
11: CVY3 |
79 |
79 |
77 |
81 |
79 |
79 |
80 |
94 |
89 |
87 |
100 |
84 |
|
12: CVY1 |
77 |
75 |
75 |
78 |
76 |
78 |
77 |
90 |
86 |
83 |
84 |
100 |
Table 5 NIP of the field sequences respects representative lineage strain
This analysis was carried out based on the alignment of 30 nucleotide sequences of the Fsp region, in which the four sequences of the isolates of this title report corresponding to Chile were included, 25 sequences of isolations from different geographical regions (America, Asia and Europe) and 1 sequence of the Measles virus Fsp as an external group (Table 3). The tree constructed segregated the four samples in the lineage America 1, including the sequence CDV/ CVy4, which previously analyzed28 was identified as part of the lineage Europe 1 (Figure 5).
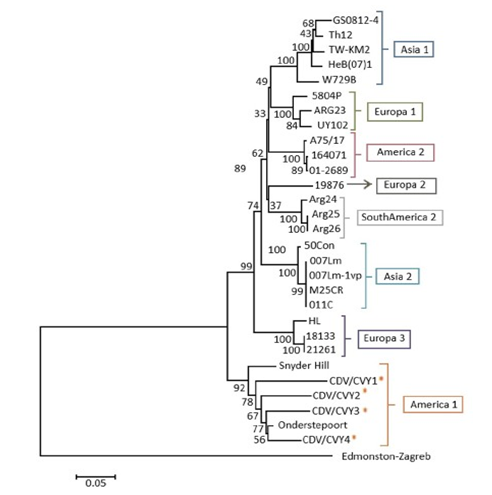
Figure 5 Evolutionary relationships of the Fsp region. Evolutionary analysis performed in the MEGA6 program using the neighbor joining method. The evolutionary distances were calculated using the p-distances method. To determine the robustness of the tree, a bootstrap of a thousand replicas was used. The analysis included 30 nucleotide sequences. The sequences analyzed in this report are marked with (*).
DC is one of the viral diseases with greater incidence and relevance worldwide in domestic and wild canines. For several decades there have been continuous outbreaks of the disease in the world, as well as a worrying expansion in the range of hosts.11,31,32 The introduction in the fifties of VVA vaccines against this virus and its extensive use have helped to keep the disease under control.8 Nevertheless, in recent decades, an increase in the incidence of this disease in canine populations throughout the world has been observed, and several episodes of the disease have also been observed in vaccinated animals.8,33 The vaccines developed to combat this disease are prepared with virus strains belonging to the lineage America 1 (Onderstepoort, Snyder Hill, Lederle, among others). In our country at least two of the eight lineages described for CDV would circulate according to the analysis of gene H: America 1 and Europe 1.28 In this study we sought to know the genomic variation of the virus through the analysis of the variable region of the F protein of CDV, using RT-PCR to amplify a fragment that contains it. This, because it has been established that the Fsp region has a genomic variability greater than that detected in the H protein (up to 27% at the amino acid level between field isolates) and consists only of 405 bp, which facilitates its amplification.23 On the other hand, the possible implications of Fsp on the function of mature F protein and in viral pathogenesis have been analyzed. Studies carried out with mutant strains in the Fsp region showed that the presence of specific mutations has drastic effects on the fusion activity of the F protein. In this way,23 generated mutants with changes in one of the two codons of start present in the Fsp, detecting a fusion activity 20 times higher in the mutants with respect to the parental viruses. In the same way, it was established that the length of the Fsp influences the activity levels of the F protein through the analysis of mutants with variable length Fsp, which revealed that the variant with the shortest Fsp (28 aa) presented a fusion activity 70 times greater than the parental protein, so they propose that this region could modulate the function of the F protein, since proteins with specific mutations or a shorter Fsp had higher fusogenic activity in vitro.23
The RT-PCR technique implemented for this work was able to amplify a fragment of around 500 bp, in the 4 positive controls and in 11 of the 22 samples analyzed (50%) and it was not amplified in the negative controls. Although these results indicate that the technique is functioning and is useful for this study, its use as a diagnostic method is not recommended because it amplifies a region that presents a large genomic variability, being more propitious, when implementing a diagnostic method, to select a conserved sequence of the genome to carry out the amplification.18 Possible explanations of why half of the samples turned out to be false-negative may be related to the amount of RNA insufficient in those samples to perform the reaction with these splitters specifically, because RNA extraction was not performed immediately after extracted the sample.28 As it could be observed in the results with samples 7 and 8, for this RT-PCR protocol it would be important to extract the viral RNA immediately after taking the patient's blood. In addition, when comparing these results with those obtained by34 it is observed, according to their results, that the samples that generated more intense bands are positive to the protocol developed in this title memory, whereas those that generate fainter bands are negative. It should be noted that one of the negative controls, both for this method and for the one carried out for the N gene, corresponded to an adult dog with all its vaccines per day, its last vaccination being ten months before the blood sample was taken. This isolated result would corroborate the idea that previous vaccinations would not cause false-positives.18,34 It is known that the viral RNA after its extraction can be very labile, being complex to work with it because it can be easily degraded by RNAs as of the skin and by temperatures higher than 4°C. In storage it can remain indefinitely active at -70° C, -192°C (liquid nitrogen) or lyophilized and remains active for about a month at -10°C.35 Considering that the RNA isolates used for this title report were extracted more than a year ago and have been stored at -20°C in the laboratory, it could be thought that the integrity of the genetic material would be lost due to the excessive storage time at a temperature Not fit. Despite this, the results show that this factor did not cause RNA degradation. Of the 11 positive samples to the RT-PCR, four were sequenced that fulfilled different parameters:
Using the BLAST program, it was confirmed that these sequences belong to CDV because all the sequences delivered by the program corresponded to the virus. The amplified sequences presented between 80 to 89% of nucleotide identity with respect to the first 100 results delivered by the program. with this, it is verified that the implemented method is specific for the detection of CDV, and that with these results of nucleotide identity the variable of the region chosen for the amplification can be evidenced when aligning the field sequences (CDV/CVY1, CDV/CVY2, CDV/CVY3 and CDV/CVY4), 107 nucleotide sites were observed that were variable among themselves, with values of nucleotide identity between 89 and 83%. These differences appear to be independent of the characteristics of the canines, since no greater homology is observed between the sequences of patients who present the same clinical signs or those who are vaccinated. There is also no greater variation in the sequence of the patient with respiratory signs, nor in the sequence of the older patient. Regarding the comparison of the sequences of the field samples with respect to the vaccines, it should be mentioned that the highest identity is achieved with the Onderstepoort strain. The above suggests that a successful vaccination alternative would be with the Nobivac® vaccine, which contains this strain. When the origin of vaccines applied to dogs that were immunized was unknown, it cannot be ruled out that a vaccine containing the Onderstepoort strain could be more or less effective against DC, considering also the study by,28 which detected the Europe 1 lineage, and, being a distinct lineage from that of vaccine strain, could be part of the reasons why vaccinated dogs acquire this disease. In the comparison between the sequences of the reference strains of the known, it was observed that the sequences of the same lineage have quite high percentages of identity among themselves (92-100%), while among the different lineages they distance themselves a little more (82-90%). For the elaboration of the phylogenetic trees, two methods were tested: "neighbor-joining" and "maximum likelihood". Two trees were built for each method with different evolutionary models and, finally, it was decided to leave the tree built by the "neighbor-joining" method with the "p-distances" model because it delivered the best values of "bootstrap".
The phylogenetic analysis revealed that the four amplified samples belong to the Latin 1 lineage, mostly related to the Onderstepoort vaccine strain. This result agrees with the comparative analysis performed on the sequences. Although, in the study by,28 the sample of canine H corresponded to the Europe 1 lineage, in this study the sequence segregated, like the other three, in the lineage America 1. This result could suggest that both regions are experiencing mutations independently in their nucleotide sequences, varying the H gene to the European lineage and the Fsp region to the American one; or that when studying a limited section of the H gene, as is the case of,28 it could generate different segregation, since only part of the gene is analyzed and not the complete gene. To elucidate this, it would be advisable to analyze the sample with both techniques to be sure that it corresponds to the indicated sample and to rule out possible confusion between the samples due to the long storage time. When the America 1 lineage was presented both in immunized and non-immunized dogs, and added to the degree of variation between the sequences of the samples collected with respect to the vaccine strain, it could be inferred that the sequences analyzed correspond to field strains belonging to This lineage, although it is not possible to completely rule out that in the case of canine H it is a reversal of the attenuated strain, due to the high percentage of identity observed (95%). To elucidate this, it would be necessary to know the strain with which the dogs were immunized. It is observed that the evolutionary process of the H and Fsp genes is equivalent, since the sequences are grouped in the same way in the phylogenetic trees, grouping in the same lineages (Asia 1, Europe 1, America 2, Europe 2, South America, Asia 2, Europe 3 and America 1). This is consistent with previous studies conducted in Asia24,25 and in Uruguay27 establishing that both genomic regions would offer the same information because they observe the same phylogenetic relationships. In addition, in27 it is mentioned that the phylogenetic relationships and the values of variability detected in the Fsp, would allow to propose a new criterion to define lineages of CDV based on this region: two isolates would belong to the same lineage if they are grouped in the same clade and present values of amino acid divergence less than 19%, while they belong to different lineages if said values are higher than 19% and are grouped in different clades.
While the lineages recognized so far are the eight mentioned by28 for the H gene, in the study conducted by27 a South American lineage is mentioned that groups three samples belonging to Argentina and that would be genetically related to the lineage Europe 2 (Wild- life) characteristic of wild carnivores, which groups isolated strains of ferrets, mink and red foxes.36 This relationship could suggest that this new lineage could come from the wildlife of South America and would be infecting domestic dogs.27 In this work, the sequences belonging to the South American lineage also segregated into a separate clade, corroborating the idea of the existence of a new lineage.
As for the rest of the research on the phylogeny of the virus carried out in South America, the Europe 1 lineage2,19,27 would be present in Uruguay and Brazil. that in Chile according to the study carried out by28 and it is postulated that the existence of this lineage in the region may be related to the importation of animals from the European continent, facilitating the spread of the virus to our continent. On the other hand, in Ecuador and Colombia, recent studies have shown the existence of a new lineage, called South America 3 in both regions, through independent studies, it is still necessary to establish whether these strains belong to the same lineage or correspond to different lineages.10,17 These results show the existence of a great variety of lineages present in ourcontinent, which is why vaccines would not be completely effective in controlling the disease.
In conclusion, the Fsp region is a good indicator to perform phylogeny of the CDV since, being a more limited region than the H gene, it would allow us to make a more rapid and accurate classification of the circulating CDV strains in each geographical region.37,38
None.
The authors declare that there is no conflict of interest.

©2021 Vera, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.