Journal of
eISSN: 2469 - 2786


Research Article Volume 5 Issue 7
Botany Department Faculty of science, Faculty of science, Tanta University, Egypt
Correspondence: Samah Abd El-Kader El-Debaiky, Botany Department, Faculty of Science, Tanta University, Egypt, Tel 00201005256803
Received: December 13, 2017 | Published: December 19, 2017
Citation: El-Debaiky SA. New record of Lecanicillium aphanocladii family: Cordycipitaceae from Egypt. J Bacteriol Mycol Open Access. 2017;5(7):430-432 DOI: 10.15406/jbmoa.2017.05.00161
New record of the entomopathogenic fungus, Lecanicillium aphanocladii (Zare & W. Gams) for the first time in Egypt. The detailed description of the cultural and morphological features of L. aphanocladii using PDA plates was exhibited in this study. In the meantime, a measurement of its microscopic structures (hypha, phialides and conidia) was carried out using ocular micrometer. The definition of L. aphanocladii was confirmed by molecular characterization by 18S rRNA gene sequences using [ITS1] and [ITS4] primers.
Keywords: lecanicillium aphanocladii, entomopathogen, molecular characterization, ITS, conidia and aphanophialides
The fungus Lecanicillium is belonging to the order Hypocreales and is described as anamorphic Cordycipitaceae. This genus is known by its ability to infect other species of fungi such as Agaricus spp., Sphaerotheca spp.1 Puccinia spp.2 On the other hand, it was recorded as entomopathogenic fungus as it can parasitize some species of insects like Mosquito larvae3-6 Bombyx mori, Trialeurodes vaporariorum. In the meantime, it is reported from leaf litter of Abelmoschus esculentus and Acacia karroo. Currently, there are 21 described species of Lecanicillium.7 In the past, this genus was described as Verticillium lecanii (Zimmerman) Viegas.
The species L. aphanocladii was described for the first time by Zare and Gams, 20018 as fungicolous fungus causing cobweb and spotting in the cultivated Agaricus mushroom. There are two synonyms (Ex-epitypes) of L. aphanocladii where it was earlier identified as:
The present study introduces a new record of L. aphanocladii in Egypt. It was discovered as a contaminant of some prepared PDA plates which stored in the laboratory at room temperature (about 26°C±2). This contamination was predicted to be due to the passage of some small insects like mites within the PDA plates. Accordingly, the present study aimed to purify the fungus and describing its cultural and morphological features in detail. The identification of the new recorded L. aphanocladii was confirmed according to molecular characterization and DNA sequencing.
Preparation of potato dextrose agar medium (PDA):
The used medium in this study was PDA9 which composed of (g/L), potato tubers, 200, D-Glucose, 20, agar, 20. The potato tubers were peeled, divided to small pieces and boiled in 1L distilled water for 1h. After boiling, the potatoes were removed by sieve and the filtrate was added to glucose and agar then the mixture volume was completed to one liter with distilled water. Chloramphenicol antibiotic (0.5 g/L) was added to prevent bacterial contamination before sterilization in autoclave at 121°C, 1.5 atm. for 15 min.
Identification of the tested fungus
Morphological characterization: The tested fungus was investigated as a laboratory contaminant on pre-prepared PDA plates. Then it was sub-cultured on new PDA plates in aseptic conditions and incubated for 5days at 26±°C. After the incubation period, the macroscopic and microscopic features of the tested fungus were recorded. The macroscopic features included the colony color and appearance, pigmentation at the bottom of the plates while the microscopic features included size and shape of conidia, conidiogenous cells (phialides) and their arrangement on the hyphae.
Molecular characterization: Molecular characterization of the fungal strain was carried out by sequencing of rRNA gene with the help of Solgent Company, Daejeon South Korea. The fungal mycelium was grown on PDA plates at 28°C for 7 days. A small portion of the mycelium was scraped and suspended in 100µl autoclaved distilled water in 2ml sterile vials and boiled at 100°C for 15 minutes. DNA was extracted and purified from the non-living fungal mycelia using SolGent purification bead. Prior to the sequencing, the ribosomal rRNA gene (rDNA) was amplified using the polymerase chain reaction (PCR) technique. Two universal fungal primers ITS1 (forward) and ITS4 (reverse) were incorporated in the reaction mixture in PCR technique.10 The sequence of each primer is: ITS1 (5' - TCC GTA GGT GAA CCT GCG G - 3'), and ITS4 (5'- TCC TCC GCT TAT TGA TAT GC -3'). The purified PCR products (amplicons) were reconfirmed using a size nucleotide marker (100 base pairs) by electrophoreses on 1% agarose gel. Then these bands were eluted and sequenced with the incorporation of dideoxynucleotides (dd NTPs) in the reaction mixture. Each sample was sequenced in the sense and antisense directions using ITS1 and ITS4 primers.10 Sequences were further analyzed using Basic Local Alignment Search Tool (BLAST) from the National Center of Biotechnology Information (NCBI) website. Phylogenetic analysis of sequences was done with the help of MegAlign (DNA Star) software version 5.05.
Identification of the tested fungus
The tested fungus was identified as L. aphanocladii based on its morphological characters which then confirmed and supported by the molecular characterization. The culture of the new recorded L. aphanocladii was deposited at the mycological center, Assuite University, Assuite, Egypt with strain number (AUMC11913).
Morphological characterization of L. aphanocladii
Macroscopic features: Colonies of L. aphanocladii were fast growing reached 3-5 cm in 4 days on PDA medium. They were characterized by their white color and high on the surface of the medium (Figure 1A). The bottom of the plates appeared with reddish reverse (Figure 1B).
Microscopic features: The phialides of L. aphanocladii were characteristic as aphanophialides which protrude solitary, paired or in cluster on the prostrate hyphae (Figure 2A, 2C & 2D). The aphanophialides were cylindrical shaped (11.5 ˟ 2.6 μ), oval shaped with small neck (3.6-4.7 ˟ 1.5-1.9 μ) or flask shaped with long tapered neck (7 ˟ 1.7 μ). Solitary conidium was observed attached to the apex of each phialide. Conidia were characteristic by different shapes; sub-globose (3.5 ˟ 2.9 μ), oval (4.9 ˟ 3.2 μ) or ellipsoidal shaped (5.7 ˟ 2.4 μ). They were generated either as phialo-conidia (Figure 2A, 2C & 2D) or directly on the hyphae (Figure 2B). These morphological characteristics of L. aphanocladii agreed the description recorded by.8
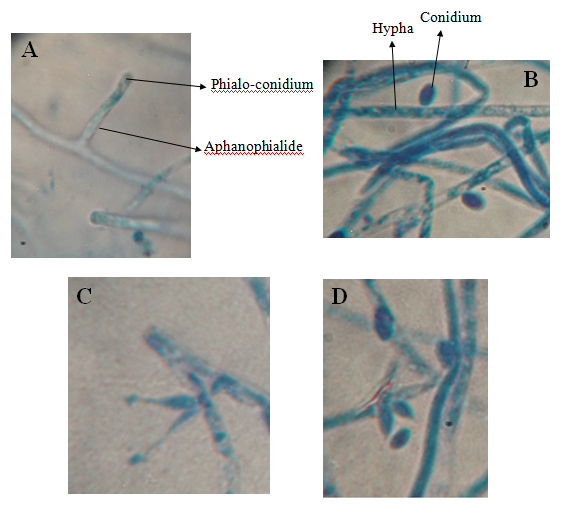
Figure 2 Microscopic structures of L. apanocladii. Cylindrical shaped aphanophialide carrying phialo-conidium (A), conidia generated directly on the hypha (B), paired aphanophialides (C) and aphanophialides in cluster (D).
Molecular characterization
The resulted data from rDNA sequencing process of the tested isolate were deposited in the GenBank with accession numbers listed in Table 1. A BLAST search for the 18S sequences of the tested isolate, L. aphanocladii (AUMC11913) indicated that it was 100 % similar to three Chinese isolates of L. aphanocladii (JX241644, JF911775 and KX530079) Table 1. The present isolate along with another recorded seven isolates of L. aphanocladii were subjected to a phylogenetic analysis using available ITS rDNA sequence data downloaded from GenBank (Table 1).
Strain No. |
Identification |
Closely Related Strains |
|||
AUMC 11913 |
Lecanicillium aphanocladii |
Strain No. |
Source/Country |
Accession No. |
Similarity (%) |
GL0836 |
Apple / China |
JX241644 |
100 |
||
HQ69 |
Lake / China |
JF911775 |
100 |
||
-------- |
Red spot of Tremella / China |
KX530079 |
100 |
||
FS13 |
Leaves of Hupezia / China |
KP689216 |
99.09 |
||
FJTL |
Tobacco / China |
KJ162356 |
99.88 |
||
DP11LA |
Leaves mining insects / Lithuania |
KC574075 |
99.66 |
||
Outgroup strain= Lecanicillium lecanii |
PA196 |
Entomopathogenic |
AF172335 |
---------- |
|
Table 1 Molecular identification of the tested fungus (AUMC11913) and percentage similarities with related strains accessed from the GenBank
The Phylogenetic tree of 18S sequences showed that my isolate clustered with L. aphanocladii isolates reported elsewhere (Figure 3). The isolate L. lecanii AF172335 was included as an out-group strain where it does not cluster with the L. aphanocladii isolate reported in this study. The resulted data of molecular and phylogenetic analyses supported the morphological identification of my isolate as L. aphanocladii.
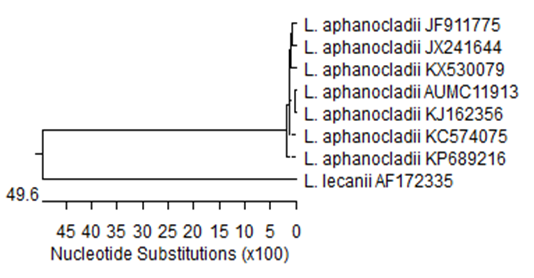
Figure 3 Phylogenetic tree of 18S sequences of the fungal strain isolated in the present study (L. aphanocladii AUMC 11913; arrowed) aligned with closely related sequences of strains accessed from the GenBank. L. = Lecanicillium, L. lecanii AF172335 is included as an out-group strain.
Later and for a long time, the fungus L. aphanocladii has been confused with the species, Aphanocladium album (Preuss) W. Gams, but the genetics studies on the compatibility among fungal species improved that the two taxa are phylogenetically quite unrelated.11,12 On the other time, a recent study revealed that there is a close relationship between L. aphanocladii and L. psalliotae based on the ITS phylogeny analysis.13
In the present study, L. aphanocladii was recorded for the first time in Egypt. The fungus was observed as a laboratory contaminant in stored PDA plates. It was identified morphologically according to its culture appearance and microscopic structures. This identification was confirmed and supported by the molecular characterization of the fungus using the 18S sequences where it shows 100 % similarities with other 6 isolates of the fungus recorded in the GenBank.
I would like to express my sincere thanks to Prof. Dr. Ahmed Moharram, mycological center, Assuit University, Egypt for his valuable assistance.
The author declares no conflict of interest.

©2017 El-Debaiky. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.