Journal of
eISSN: 2469 - 2786


Research Article Volume 4 Issue 6
Division of Plant Pathology, Plant Protection Research Institute (PPRI), Viet Nam
Correspondence: Le Thi Thanh Tam, Division of Plant Pathology, Plant Protection Research Institute (PPRI), Vietnam Academy of Agricultural Science (VAAS), Duc Thang, Bac Tu Liem, Hanoi, Viet Nam
Received: February 02, 2017 | Published: May 18, 2017
Citation: Tam LTT. Identification powdery mildew erysiphe quercicola damaging on mango in Hanoi, Vietnam. J Bacteriol Mycol Open Access. 2017;4(6):175-179. DOI: 10.15406/jbmoa.2017.04.00111
Powder mildews, caused by the Erysiphales, which consist of various different tribes, genera and species, are popular diseases causing severe damage on many economically important crops in Vietnam. The identification is important for biocontrol as well as fullfilling gaps in knowledge of the world about their distributions and epidemics. In current study, Erysiphe quercicola has been discovered in Vietnam for the first time as causal agent of powdery mildew disease on mango (Mangifera indica) based on both molecular and morphological classifications and pathogenicity test as well. Phylogenetic analysis using ITS sequences clearly demonstrated that this species in Vietnam belongs to E. quercicola group with strong bootstrap supports of 99% in both MP, PAUP 4.0 and ML, MEGA7 analyses. The 28S ribosomal DNA sequence analysis for better elucidation its phylogeny and taxonomic structure also has had similar result.
Keywords: erysiphe quercicola, its, 28S, phylogeny, powdery mildew, mango
During the survey and studying of powdery mildews from 2012 to 2015; several powdery mildew species have been identified first time in Vietnam. One of the most important Erysiphales that has been found in Vietnam is belonged to Erysiphe genus damagingon mango (Mangifera indica). This species causes early falling of mango flowers and young fruits; resulting in approximately yield loss of 92% in mango fruit production in several northern localities in Vietnam. This species was first described as Oidium species; O. mangifera on M. indica by Berthet (1914).1 Then this species was re-identified as an anamorph of Erysiphe quercicola by Takamatsu et al.2 and Braun & Cook1 by mostly morphological characteristics of sexual stage (teleomorph) or molecular phylogenetic analysis. However; this teleomorph has been not found yet in Vietnam so that it obtained morphological descriptions are not adequate to distinctly delimit the species. Therefore; in this study; phylogenetic analyses based on the ITS or 28S rDNA sequences of the powdery mildews belonging to the genus Erysiphe from specimens sampled on mango were carried to elucidate their taxonomy.
Light microscopy of fresh material for morphological observation
Hyphae; conidiophores and conidia of fresh materials were stripped off the leaf surfaces with clear adhesive tape; mounted on a microscope slide with the fungal mycelium uppermost; and examined by using light microscopy with phase contrast 40 X objectives. The following information was noted during the examination of the fresh specimens: size and shape of conidia; presence or absence of fibrosin bodies; characteristics of the conidiophores; e.g.; size and shape of foot cell; shape and position of hyphal appressoria on germ tubes of conidia. One hundred conidia were measured for each specimen examined. Thirty foot-cells were measured for each specimens. Observation of conidial germ tubes was carried out using the method of Hirata.3
DNA extraction and PCR amplification
Whole cell DNA was isolated from fresh fungal specimens or dried plant (or herbarium) specimens by purification genomic DNA from conidia via D Neasy Blood and Tissue Kit (QIAGEN). The ITS region of the nuclear rRNA including the 3’- end of the 18S (small subunit) rRNA gene;the 5.8S rRNA gene; and 5’-end of the 28S (large subunit) rRNA gene were amplified by polymerase chain reaction (PCR) with the primer pairs listed in Table 1 and Figure 1 below.
The ITS rRNA gene were amplified by PCR with the primer pairs as listed in Table 1. The first PCR products were used for the templates of the second PCR using the same primer set. PCR reactions were conducted with TaKaRa Taq DNA polymerase (TaKaRa; Tokyo) under the following thermal cycling conditions in a PCR thermal cycler SP (Takara; Kyoto; Japan): an initial step for denaturing at 94˚C for 3 min; thermo cycling for 30 cycles that each cycle consisted of 30s at 94˚C; followed by 30s at 58˚C for annealing for primer pairs HF1/HR4;4 and a final extension cycle at 72˚C for 10 min. The 1st PCR were conducted in 20µl total including Taq PCR Master Mix (TaKaRa; Tokyo) 10µl; 1.0µl each forward and reversed primer (concentration 20 mM); dd H2O 4.0µl; and gDNA powdery mildews fungi 5.0µl. The 2nd PCR reactions; were conducted in 30µl total including 20µl Taq PCR Master Mix (TaKaRa; Tokyo); 1.0µl each primer (concentration 20µM); dd H2O 5.0µl;and 3.0µl of the first PCR product.
Order No. |
Primer Pairs |
Sequence (5’- 3’) |
PCR/Sequencing |
Reference |
1 |
HF1 |
GGATCCTCGTAACAAGGTTTCCGTAG |
PCR/ |
|
HR4 |
CTGCAGCTCCGCTTATTGATATGCTT |
Sequencing ITS |
||
2 |
NL1 |
AGTAACGGCGAGTGAAGCGG |
Sequencing |
|
NLP2 |
GGTCCCAACAGCTATGCTCT |
28S |
Table 1 List of primer pairs using for PCR or sequencing ITS rDNA or 28 rDNA of powdery mildew fungal species (PPRI 2014)
The 28S rRNA gene were amplified by PCR with the primer pairs as listed in Table 1. PCR reactions were conducted with TaKaRa Taq DNA polymerase (TaKaRa; Tokyo) under the following thermal cycling conditions in a PCR thermal cycler SP (Takara; Kyoto; Japan): an initial step for denaturing at 94˚C for 3 min; thermo cycling for 30 cycles that each cycle consisted of 30s at 94˚C; followed by 30s at 50˚C for annealing; and 54s at 72˚C for extension; and a final extension cycle at 72˚C for 10 min. The 1st PCR were conducted in 20µl total including Taq PCR Master Mix (TaKaRa; Tokyo) 10µl; 1.0µl each forward and reversed primer (concentration 20µM); and gDNA powdery mildews fungi 9.0µl. 2nd PCR reactions were conducted in 30µl total including 20µl Taq PCR Master Mix; 1.0µl each primer (concentration 20µM); dd H2O 2.0µl and 6.0µl of the first PCR product.
The PCR amplicons were separated by electrophoresis on 2% agarose gel in 1% TAE buffer; staining by being immerged in 1% TAE solution with Green safe or Ethidium Bromide for 30mins. The desired band was visualized under long-wavelength ultraviolet light and cut out of the gel. Purification of the DNA fragment was performed utilizing the QIAquick Gel Extraction Kit (QIAGEN); as described by the manufacturer’s protocol.
Sequencing
For ITS and 28S rDNA sequencing; both strands of the amplicons were sequenced using the primer sets as listed in Table 1. The nucleotide fragments of the second PCR products after purification were sent to First Base Co. (Malaysia/Singapore) or Macrogen Co. (Korea) for sequencing.
Phylogenetic analysis
Sequence assembled by using DNAstar Lasergene 11 Core Suite software (http://www.dnastar.com/t-allproducts.aspx). Then;using the ClustalX package5 to align the sequences determined in this study with sequences of the powdery mildew fungi obtained from DDBJ database which have high identity after using BLAST for searching. The alignment was then visually refined in MEGA 7. Phylogenetic trees were obtained from the data using the maximum parsimorny (MP) and maximum likelihood (ML) analyses. GenBank codes for ITS and 28S sequences in this study as well as extracted from Database were represented along with taxon name in phylogenic trees shown in result section.
MP analysis was performed in PAUP 4.0 b106 with the heuristic search option using the tree bisection-reconstruction (TBR) algorithm. The strength of the internal branches of the resulting trees was tested with bootstrap (BS) analysis;7 using 1000 replications with the stepwise addition option set to simple and a maximum tree number of 100.
The ML analysis was done using MEGA 7 version for big datasets.8 The trees with bootstrap value supports were obtained by running 1000 replication.
Inoculation test
Each inoculation was applied on ten 7-days old-leaves and replicated three times; in chamber room conditions: temperature 25˚C; relative humidity 90%;light intensity 12000lux as method of.9 Using ANOVA and Duncan test in SAT/STAT 9.1.3 Portable for statistically analyzing.
Morphological study
The morphological characteristics of the powdery mildew fungi were analyzed in this study shown in Figure 2.
Conidiophores have foot cell measured from 37.5 to 40.0 μm in length and from 8.8 to 12.5µm in wide. Conidia produced singly; ovoid; without fibrosin bodies; and measured from 32.5 to 42.5μm in length and from 17.5 to 22.5μm in wide with a length/width ratio from (1.4-) 1.6 to 2.3(-2.6). Appressoria on mycelium were lobed or multilobed. Conidia germinated with Extensitubus subtype of Pseudoidium type germ tubes. Therefore; this powdery mildew which is damaging on mango in Vietnam would belong to the genus Erysipheaccording to Braun & Cook.1 However; due to the absence of chasmothecia; it is very difficult to identify species for this powdery mildew without using molecular techniques.
Molecular phylogenetic study
ITS phylogenic analysis: Five ITS sequences of powdery mildew on mango which have already obtained in this study including isolates M1-M5 with Genbank accession Nos. KM260685-KM260690 were aligned with 28 sequences representing Microsphaeralineage of the genus Erysiphe and one; Erysiphe glycines AB015934; from the Uncinula lineage; used as outgroup taxa extracted from GenBank. The alignment data matrix consisted of 34 taxa and 576 characters; in which 434 (75.35%) characters are constant and 62 (10.76%) variable characters are parsimony-uninformative. This parsimony analysis using PAUP* generated best tree; No. 1; described as Figure 3 with boostrap value (BP). The tree topology of the NJ tree with BP was similar to the MP tree (Supplemental Figure 1).
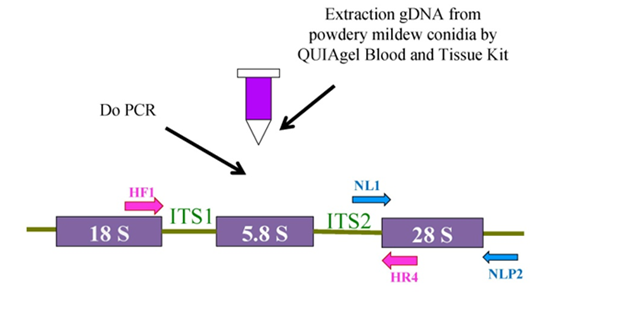
Figure 1 Scheme for using primers pairs to PCR or sequence ITS or 28S rDNA of powdery mildew fungus from conidia (PPRI 2004).
The phylogenetic tree based on analysis ITS rDNA sequences in Figure 3 clearly indicated that powdery mildew on mango (Mangifera indica) in Vietnam belonged to E. quercicola clade with strong bootstrap support (both 99% in MP and in ML analysis (Supplemental Figure 1). Meanwhile; in phylogenetic analysis carried out by Takamatsu et al.2 powdery mildew fungi on belonging to E. quercicola clade with 99%BP values support. Especially; in our study; powdery mildews on C. reticulate (mandarine); M. indica (mango); H. brasiliensis (para rubber tree) in Vietnam; all belonged to E. quercicola clade.
28S phylogenic analysis: Because the identity is so high among ITS regions of E. quercicola species so that 28S rDNA phylogenic analysis should be carried to better elucidate their phylogeny and taxonomic structure. Five 28S sequences of powdery mildew on mango which have already obtained in this study including isolates M1-M5 with Genbank accession Nos. KM260667-KM260672 were aligned with 34 sequences representing Microsphaera lineage of the genus Erysiphe and one; Erysiphe australiana AB022407; from the Uncinula lineage; used as outgroup taxa extracted from GenBank. The alignment data matrix consisted of 40 taxa and 730 characters; in which 311 (42.60%) characters are constant and 45 (6.16%) variable characters are parsimony-uninformative. This parsimony analysis using PAUP* generated best tree described as Figure 4 with boostrap value (BP). The tree topology of the NJ tree with BP was similar to the MP tree (Supplemental Figure 2).


Figure 2 Some morphological characteristics of anamorph stage of powdery mildew fungi damaging mango in Vietnam. A-B: Symptoms of powdery mildew fungi on leaves of mango; C: Conidia germination with Pseudoidium type and Extensitubus sub-type of conidia of powdery mildew fungi in mango. Bar for measurement: C: 15µm. (PPRI 2014).

Figure 3 Phylogenetic analysis of the ITS rRNA region for 34 sequences from the genus Erysiphe in which 33 ones from the Microsphaeralineage and one, Erysiphe glycines AB015934, from the Uncinulalineage, used as outgroup taxa. The tree was obtained by a heuristic search employing the random stepwise addition option of PAUP.6 Gaps were treated as missing data. Percentage BS support values (1K replications; >50 %) are shown on branches. ITS rDNA sequences of powdery mildews on mango (Mangifera indica) determined in this study are shown in boldface along with the ones on mandarine (Citrus reticulate) and para-rubber tree (Hevea brasiliensis) which were defined in Vietnam in previous reports respectively by Tam et al.13,14

Figure 4 A single MP tree based on the sequences of the 28S rDNA from 40 isolates of the genus Erysiphe in which 39 ones from the Microsphaera lineage and one, Erysiphe australiana AB022407, from the Uncinula lineage, used as outgroup taxa. The tree was obtained by a heuristic search employing the random stepwise addition option of PAUP.6 Gaps were treated as missing data. Percentage BS support values (1K replications; >50 %) are shown on branches. 28S rDNA sequences of powdery mildew on mango (Mangifera indica) determined in this study are shown in boldface along with the ones on mandarine (Citrus reticulate) and para-rubber tree (Hevea brasiliensis) which were defined in Vietnam in previous reports respectively by Tam et al.13,14
Once again; the result of 28S rDNA phylogenetic study indicated that the powdery mildew mango in Vietnam belong to E. quercicola clade with bootstrap value support 58.1% and 69% in MP and ML analysis; respectively.
Inoculation test
Pathogenicity for E. quercicola on mango was confirmed through inoculation tests by gently sweeping conidia by pen brush from diseased mango leaves onto 7-10 days-old fresh mango leaves. These ten inoculated mango leaves were kept on floating on the water surface in ten plates separately. Ten plates with ten non-inoculated fresh mango leaves were used as controls. All was kept in chamber room with optional conditions including temperature 25˚C; relative humidity 90%; light intensity 12000lux as method of Pab et al.9 Inoculated leaves developed significant symptoms almost after 3 days; whereas the control leaves remained symptomless. The E. quercicola present on the inoculated leaves was morphologically identical to that observed on the diseased original leaves; with same PCR result; fulfilling Koch’s postulates.
At present; in the world; more than 700 powdery mildews species are known; but cross-inoculation studies and molecular sequence analyses are only known for a few of them. In addition; it is necessary to carry out surveys of regions where little is known about powdery mildews with the object of publishing local checklists; host ranges and distribution data. Moreover; molecular techniques should be improved to prove the identity of anamorphic states; above all in case of new introductions and in regions such as the tropics and subtropics where they often have little inclination to form the teleomorph.10-15
Therefore; in our study; first time; it is pronounced to find out species of powdery mildews fungi Erysiphe quercicola that damaged severely mango flower and fruit in Gia Lam; Hanoi; Vietnam by both morphological and molecular classifications and pathology test as well.
Powdery mildew fungi damaged on mango in Gia Lam; Hanoi; Vietnam has been defined as Erysiphe quercicola.
None.
The author declares no conflict of interest.

©2017 Tam. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.