Journal of
eISSN: 2469 - 2786


Research Article Volume 8 Issue 1
Department of Medical Microbiology, Dicle University, Turkey
Correspondence: Nida ÖZCAN, Dicle University Faculty of Medicine, Department of Medical Microbiology, Diyarbakır, Turkey
Received: December 27, 2019 | Published: January 31, 2020
Citation: ÖZCAN N, YAKUT S, GENİŞEL N, et al. Identification and antimicrobial susceptibilities of Enterobacter species isolated from clinical specimen in southeastern Turkey from 2015 to 2017. J Bacteriol Mycol Open Access. 2020;8(1):175-178 DOI: 10.15406/jbmoa.2020.08.00265
Background/aim: Enterobacter species often colonise human gastrointestinal tract, causing various opportunistic infections. Enterobacter cloacae and Enterobacter asburiae are the most frequently isolated Enterobacter species. The aim of this research was to investigate antimicrobial resistance among Enterobacter spp. strains isolated from patients in a tertiary hospital of Southeastern Turkey. There are few publications on antibiotic resistance of Enterobacter species.
Materials and methods: This retrospective study included Enterobacter spp. strains isolated from clinical specimen sent from Dicle University Hospital clinics from 2015 to 2017. The isolates to be considered as infection agents were identified by MALDI-TOF mass spectrometry. The antimicrobial susceptibility test (AST) was carried out by semi-automated microbiology system and evaluated according to EUCAST v.8.0 criteria.
Results: A total of 296 (93 in 2015, 96 in 2016, and 107 in 2017) Enterobacter spp. strains was were isolated over three-years period. The most frequently identified species was E. cloacae (240 strains, 81.1%). The highest resistance was found against aztreonam (41.8%) and ceftazidim (41.9%) while lowest resistance was against amikacin, meropenem and ciprofloxacin. Conclusion: Amikacin and meropenem were the most effective antibiotics against E. cloacae. The resistance rates of other Enterobacter strains other than E. cloacae varied according to years and species.
Keywords: Enterobacter cloacae, antibiotic resistance, amikacin, meropenem
Enterobacter species are members of human gut microbiota. These facultative anaerobe Gram negative rods may cause opportunistic infections especially in immunocompromised patients.1 Hoffman et al. described E.cloacae, E.asburiae, E.hormaechei and E.kobei species as ’’E.cloacae complex’’ in 2005.2–5 Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumanni, Pseudomonas aeruginosa ve Enterobacter spp. are important infectious agents which are briefly named as “ESKAPE” pathogens. Enterobacter species may have multiple drug resistance with porine loss, efflux system activation, AmpC cephalosporinase and metallo-betalactamase enzyme systems.6 In particular, carbapenem resistant isolates with blaNDM1 and blaKPC enzymes have been reported to cause serious nosocomial outbreaks in different countries.5,7–9 The most frequently isolated Enterobacter species have been reported as E.cloacae, E.aerogenes and E.asburiae worldwide.5 Microscobic examination of the specimen is important to determine whether bacteria isolated in culture are causative agents of infection. Number of leucocytes indicates whether the culture growing bacteria were infectious agents or colonizers.10,11 The Q score assessment indicates the number of polymorphonuclear cells (PMN) with positive values and the number of squamous epithelial cells (SEC) with negative values directly seen in Gram-stained smears of wounds.12 Matrix-assisted laser desorption/ionization- time of flight- mass spectrometry (MALDI-TOF-MS) has been used as a tool for bacterial identification in the last decades. The system provides fast and easy identification of bacteria that grow in culture.4,13 Articles on Enterobacter species alone are quite limited in our country. Current study aims to identify and determine antibiotic susceptibility of Enterobacter species isolated from various clinical samples in our laboratory between 2015-2017.
The approval form of this retrospective study was obtained from Dicle University Ethics Committee (No: 124) in March 2019. Enterobacter strains isolated from different specimen sent from clinics of Dicle University Hospital between January 2015 and December 2017 were included in the study. Cerebrospinal fluid (CSF), pleural fluid, peripheral and catheter blood samples were taken into Bactec Plus aerobic / F or BD Bactec Peds Plus / F bottles and incubated in BACTEC FX (Becton Dickinson, USA) system. Bottles with bacterial growth signal were subcultured on solid media. For the respiratory tract and wound samples, Bartlett and Q scoring methods of microscobic examination were used to determine whether the isolate was an infectious agent.10 The same bacteria grown in both catheter and peripheral blood samples was taken as causative agent of catheter-related bloodstream infections. Bacterial growth of the catheter samples with no growth in the peripheral blood culture was evaluated as contamination or colonization.14 The causative agents according to Gram staining and culture growing profiles were identified with MALDI-TOF mass spectrometry by Maldi Biotyper (Bruker, U.S.A). Antibiotic susceptibility tests (AST) were performed with Phoenix 100 (Becton Dickinson, U.S.A) automated system by using Phoenix UNMIC-401 and NMIC-400 ID panels respectively for urine and other samples. EUCAST v.8.0 criteria were used for the evaluation of antimicrobial susceptibility tests.15
A total of 296 (93 in 2015, 96 in 2016 and 107 in 2017) Enterobacter spp. strains were isolated over three years’ time. About 72% (n=213) of the bacteria were isolated from clinical or intensive care patients while 28% (n=83) were isolated from outpatients. 153 (51.7%) of the patients were male and 143 (48.3%) were female. The most common isolates were urine (138 specimen, 46.6%), blood (44 specimen, 14.9%) and wound (35 specimen, 11.8%). Tracheal aspirate (23 specimen, 7.8%), tissue and abscess (17 specimen, 5.7%), catheter (7 samples, 2.4%), sputum (6 samples, 2%) and other samples (pleural, peritoneal, synovial and cerebrospinal fluids, bile, ear drainage, bronchoalveolar lavage; 26 samples, 8.8%) were also evaluated. Microscopic examination results were taken into consideration when evaluating culture results in the majority of samples. Bacterial growth in samples with abundant leukocytes and no epithelium was evaluated as the causative agent of infection (Figure 1). The most frequently identified species were E.cloacae (240 strains, 81.1%) and E.asburiae (29 strains, 9.8%) while 9 species (3%) could not be identified up to species level (Figure 2). Amikacin was found to be the most effective antibiotic against all Enterobacter isolates. Resistance rates to amikacin was found to be 5.1% among E. cloacae and 1.9% among other Enterobacter species. Gentamicin resistance rates were 24.6% among E.cloacae and 5.7% among other species. Meropenem resistance rates were found to be 10.3% and 16.4% among E.cloacae and other species, respectively. Approximately one third of all isolates were resistant to beta-lactam antibiotics (ceftazidime, cefixime and aztreonam). Ciprofloxacin resistance was found to be 21.2% and 11.1% among E. cloacae and other Enterobacter strains, respectively. Approximately one third of all isolates were resistant to beta-lactam antibiotics (ceftazidime, cefepime and aztreonam). Trimethoprim-sulfamethoxazole resistance rates among E. cloacae and other species were as 31.1%, and 10.9%, respectively (Table 1).

Figure 1 Gram staining image of a tracheal aspirate sample (with x1000 magnification) The bacteria was determined as infectious agent due to leucocytes with polymorphic nuclei.
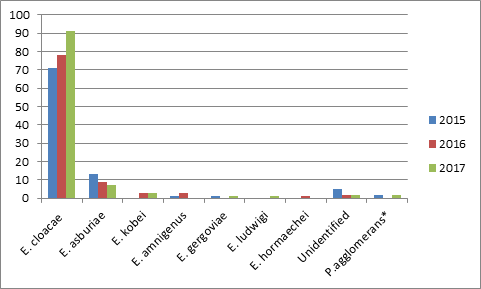
Figure 2 Distribution of Enterobacter spp. strains isolated from clinical specimen from 2015 to 2017.
*Pantoea agglomerans
| Species | E.cloacae | Other species* | Total |
| Antibiotics | |||
| Gentamicin | 58/236(24.6) | 3/53(5.7) | 61/289(21.1) |
| Amikacin | 12/237(5.1) | 1/53(1.9) | 13/290(4.5) |
| Ceftazidime | 111/233(47.8) | 8/51(15.7) | 119/284(41.9) |
| Cefepime | 97/228(42.5) | 7/54(13) | 114/330(40.4) |
| Aztreonam | 107/229(46.8) | 10/51(19.6) | 117/280(41.8) |
| Meropenem | 24/234(10.3) | 9/55(16.4) | 33/289(11.4) |
| SXT** | 74/238(31.1) | 6/55(10.9) | 80/293(27.3) |
| Ciprofloxacin | 50/236(21.2) | 6/54(11.1) | 56/341(19.3) |
Antimicrobial susceptibility test (AST) results of Enterobacter spp. strains isolated between 2015- 2017 [number of resistant strains/ number of total strains (resistance rate%)]
*:E. asburiae, E. kobei, E. amnigenus, E. gergoviae, E, ludwigi, E. hormaechei,
agglomerans ve unidentified species
**:Trimetoprim/sulfametoxazole
Enterobacter species frequently caused nosocomial infections in recent decades. Researchers reported carbapenem-resistant Enterobacter species as serious nosocomial agents with a high mortality rate. Hong et al.1 reported 51 colistin resistant strains among 213 E. cloacae isolates in Korea. Carbapenem resistant E. cloacae strains were reported by Jin et al.1,4,5,7 & Öğünç et al.16 found the resistance rates of imipenem, cefepime and amikacin as 2%, 9% and 31%, respectively, among 45 Enterobacter strains (E. cloacae and 11 E. aerogenes). İmipenem and cefepime were the most effective antibiotics while amikacin resistance was relatively high in that study. In the same study cefotaxime and ceftazidime resistance were found as 67% and 64%, respectively.16 It was found that 41.9 % of total isolates were resistant to ceftazidime and 40.4% were resistant to cefepime. In a surveillance study, Aksaray et al.17 reported that among 50 Enterobacter strains isolated as nosocomial agents 68% of strains were resistant to ceftazidime, 28% to cefepime and 66% to cefotaxime.17 In the same study, imipenem was found to be the most effective antibiotic with a resistance rate of 4%, followed by ciprofloxacin, cefepime and amikacin with 8%, 28%, 32% resistance rates, respectively.17 Kaleli et al.18 evaluated antimicrobial resistance of 109 Enterobacter strains; 15% of the strains were resistant to amikacin, while resistance to ceftazidime, ceftriaxone, cefaclor and cephalotine were found as 47%, 50%, 72% and 83% , respectively.18 Yazıcı et al.19 obtained the lowest resistance against amikacin (2.4%) followed by imipenem (2.4%) and ciprofloxacin (2.4%) in 120 Enterobacter isolates obtained from 42 outpatients and 78 inpatients.19 The antibiotics resistance rates of Enterobacter strains isolated from Dicle University hospital clinics between 2005-2007 were determined according to the Clinical & Laboratory Standards Institute (CLSI) criteria. Meropenem and amikacin were found to be the most effective antibiotics for E.aerogenes and E.cloacae strains.20 The resistance rates of both meropenem and amikacin among Enterobacter strains isolated between 2005-2007 were reported as %3. In current study the resistance rate for the meropenem and amikacin were 11.4% and 4.5%, respectively. Resistance to ciprofloxacin was reported as 27% among Enterobacter strains isolated by by Öğünç et al.16 Ciprofloxacin resistance among Enterobacter isolates were reported as 11.5% in inpatients and 4.8% in outpatients by Yazıcı et al.19 Ciprofloxacin resistance was reported as 15% by Kaleli et al.18 in 2001, and as 19% by Gülhan et al.18,20 In our study, it was found to be 19.3%, consistent with previous studies. Multi-drug resistant Enterobacter isolates have been reported to be a worldwide problem. Determination of drug resistance mechanisms are indicated to be important for treatment especially in carbapenemase producing isolates.7,8 Tarumoto et al.21 identified a blaIMI-1 secreting colistin-heteroresistant E.cloacae in an article published in 2018. The strains were reported to be resistant to cephalosporins (except fourth generation), carbapenems, levofloxacin and aminoglycosides.21
Kyaung Hong et al. investigated antibiotics resistance in colistin-resistant Enterobacter spp strains in their articles in 2018. Among 51 colistin-resistant E. cloacae isolates; meropenem, gentamicin, cefepime, ciprofloxacin, ceftazidime and trimethoprim / sulfomethoxazole resistance rates were found as 2%(1 isolate), 2%(1 isolate), 9.8%(5 isolates), 19.6% (10 isolates), 47.1%(24 isolates) and 52.9(27 isolates), respectively.1 Among 6 colistin-resistant E. aerogenes isolates; meropenem, cefepime, gentamicin, ciprofloxacin, ceftazidime and trimethoprim/sulfomethoxazole resistant rates were 16.7% (1 isolate), 16.7% (1 isolate), 50%(3 isolates), 50%(3 isolates), 83.3%(5 isolates) and 83.3%(5 isolates), respectively.1 In two articles published in 2016, researchers reported that colistin has failed in some treatments due to colistin resistance; E.cloacae and E.aerogenes species as opportunistic pathogens that can cause serious problems for human health.22,23 In a study conducted in Brazil in 2018, the researchers evaluated the resistance profiles of 14 Enterobacter isolates ( 8 E.cloacae and 6 E.aerogenes) to different antibiotics. All strains were resistant to sulfonamide, 12 (85.7%) strains to cefaclor and nitrofurantoine, 11 (78.6%) strains to doxycycline, 6 (42.9%) strains to ertapeneme, 5 (35.7%) strains to ceftazidime, cefepime and ceftriaxona, 4 (28.6%) strains were found to be resistant to amikacin, gentamicin, tobramycin, ofloxacin, cefotaxime and aztreonama, 3 (21.4%) strains were resistant to ciprofloxacin, levofloxacin and tetracycline, 2 (14.3%) strains to meropenem and 1 (7.1%) to chloramphenicole.9
Amikacin, an aminoglycoside, was an effective antibiotic against all Enterobacter isolates and the resistance rate of meropenem was higher in other Enterobacter species compared to E. cloacae strains. In this study, it was emphasized that microscopic examination contributed to the evaluation of culture results. It was also emphasized that mass spectrometry stands as a novel and current bacterial identification method.
None.
None.
The author declares no conflict of interest.

©2020 ÖZCAN, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.