Journal of
eISSN: 2469 - 2786


Research Article Volume 5 Issue 1
1Department of Zoology, Chikiti College, India
2Department of Microbiology, National Centre for Disease Control, India
3Department of Molecular Biosensor, CSIR-Institute of Genomics and Integrative Biology, India
Correspondence: Kumar Dash, Department of Zoology, Chikiti College, Chikiti, Ganjam, Odisha-761010, India, Tel 9.19439E+11
Received: May 12, 2017 | Published: June 29, 2017
Citation: Dash SK, Khare S, Kumar A. Human brain damage detection using porA marker. J Bacteriol Mycol Open Access. 2017;5(1):211-213. DOI: 10.15406/jbmoa.2017.05.00118
Human brain damage (meningitis) caused by Neisseria meningitidis is generally diagnosed from patient cerebrospinal fluid through microscopy, immunological assays, biochemical methods, PCR, microarray or biosensors. All these methods are expensive, time consuming or non-confirmatory. A quick PCR based method was developed for detection of human brain damage using specific primers based amplification of PorA gene partial sequence (211bp). The PorA gene amplicon can be used as a marker for detection of about 2mg of genomic DNA (9.36x104 colony forming unit) of N. meningitidis in 80min. This is the least time reported so far for confirmation of the disease. The cross reactivity of the marker was checked with other possible organisms present in cerebrospinal fluid. A comparative study of the available methods with the PorA as marker was done using 30 meningitis suspected patient samples.
Keywords: diagnosis, genetic marker, marker, meningitis, neisseria meningitidis, PorA gene
CFU, colony forming unit; CSF, cerebrospinal fluid; DNA, deoxyribonucleic acid; EDTA, ethylenediaminetetraacetic acid; G-DNA, genomic DNA; opc, opacity associated; PCR, polymerase chain reaction; PorA, porin protein a; RT-PCR, real time-PCR; TE, 10mM Tris and 1mM EDTA; VR, variable region
Human brain bacterial meningitis causes inflammation of the meninges in the brain and spinal cord of the patients leading damage of the brain or even death.1 The disease spreads very rapidly among people in contact leading to endemic or epidemic outbreak. The usual methods of detection of human brain damage (meningitis) includes biochemical tests,2 microscopy,3 latex agglutination test,4 PCR,5,6 RT-PCR,7 microarray,8,9 and biosensors.10-12 All these methods suffer from one or other limitations therefore, a quick and specific method is required for the detection of the disease to prevent the outbreak and early treatment of the patient. Kotilainen et al.13 detected bacterial meningitis through broad-range PCR of genes coding for 16s and 23s RNA of N. meningitidis but was marked down because of its long proteolysis step. The multiplex PCR used by Seward and Towner14 and Mohamed et al.15 involved purification of RNA and synthesis of cDNA which was time consuming. Taha16 and Lewis & Clarke17 detected sero groups of N. meningitides by crgA and siaD gene sequence analysis, which was time consuming and complicated. Baethgen et al.18 and Bronska et al.19 used purified G-DNA of N. meningitidis for PCR. Fraisier et al.20 reported a multiple gene based multiplex PCR, which was complicated and expensive. Boving et al.21 reported an eight-plex PCR for simultaneous detection of N. meningitidis, S. pneumoniae, Escherichia coli, Streptococcus aureus, Listeria monocytogenes, Streptococcus agalactiae, herpes simplex virus (types 1, 2), and varicella-zoster virus. The PCR product was detected through array and microsphere coupling method. Recently, Kumar et al.22 reported 304 bp of opc gene as a genetic marker for detection of bacterial meningitis.
The class 1 outer membrane porin protein, PorA belonging to the ST-41/ST-44 complex lineage III, is expressed by almost all meningococcal strains.23 The PorA protein consists of eight surface-exposed loops, with loops 1 and 4 each containing one variable region, VR1 and VR2, respectively.24 The antigenic variation among PorA proteins is the basis of sero grouping of N. meningitidis. The monoclonal antibodies against serosubtype-specific epitopes on PorA, exert protection against N. meningitidis mediated human brain meningitis. Therefore, National Institute of Public Health and the Environment, Netherland have developed a vaccine against N. meningitidis (serogroup B) targeting PorA as an antigen.25 Here we report a quick diagnosis of the human brain damage caused by N. meningitidis by using PorAgene as marker.
Materials
The PCR chemicals, Taq polymerase and agarose were purchased from Bangalore GeNei. EDTA was obtained from Qualigens. Tris (hydroxymethyl) aminomethane was procured from Sigma-Aldrich. DNA purification kit was purchased from Biochem Life Sciences. Primers were synthesized from GCC Biotech. The PCR purification GFX column was obtained from Biochem life science, India. N. meningitidis culture and patient cerebrospinal fluid samples were obtained from NCDC, Delhi.
PCR Amplification
Patient CSF (0.5 ml) was taken in an eppendorf and centrifuged at 16,000xg for 1 min. The supernatant was discarded and the pellet was suspended in 25µl of PCR mix containing 1X PCR buffer (1.5 mM MgCl2, 0.01 M Tris-HCl, 0.05 M KCl, 0.01% gelatin, pH 8.3), 0.4 mM dNTP mix (0.1 mM of each nucleotide), 0.4µM of each forward and reverse primers of PorA gene of N. meningitidis, 0.75U of Taq polymerase and de-ionized water. Initially, a gradient PCR was carried out in a MJ Mini (Bio-Rad) thermal cycler with the following steps: 95˚C for 5 min followed by 30 cycles of denaturation at 95˚C for 4 s, gradient annealing temperature at of 51.0, 52.2, 54.4, 57.3, 60.9, 63.8, 65.9 and 67˚C for 5 s, extension at 72˚C for 1 s and final extension of 3 min at 72˚C. The PCR was carried out at an annealing temperature of 54.4˚C for 5 s keeping other parameters same and the amplicon was analyzed in 1.5% agarose gel electrophoresis. The PCR amplicon was purified using GFX column, and sequenced at GCC Biotech.
For determination of the sensitivity, serial dilutions of N. meningitidis cells in 0.5 ml of patient CSF were taken and centrifuged at 16,000xg for 1 min. The upper viscous CSF was discarded and the pellet was suspended in 0.5 ml of TE buffer, pH 8. The bacterial suspension was heated at 95˚C for 5 min and centrifuged at 16,000xg for 1 min. The amount of the G-DNA was quantified from the supernatant of each dilution. In a second set of experiment, the N. meningitidis dilutions were plated in triplicate on Mueller-Hinton agar and grown at 37˚C for 18 h in CO2 rich environment. The average CFU count of N. meningitidis was calculated for different cell dilutions. In the third set of experiment, serial dilution of N. meningitdis cells in 0.5 ml of CSF were taken and centrifuged at 16,000xg for 1 min. The upper supernatant was discarded and the pellet was suspended in 25µl of PCR mix and the PCR was carried out as described above. The PCR amplicons were detected in 1.5 % agarose gel to determine the minimum number of N. meningitidisCFU required for PCR.
The gradient annealing temperature PCR showed comparatively less PCR yield at 51 and 52.2˚C while the amount of PCR product remained almost same from 54.4-67˚C (Figure 1A). Therefore, 54.4˚C was chosen as standard annealing temperature for the PCR amplification. The PCR product 211 bp of PorA gene of N. meningitides Figure 1B was purified using GFX column and sequenced at TCGA (Figure 1C). APCR, for quick detection of N. meningitidisin 80 min (including gel electrophoresis) directly from patient CSF was developed. The diagnosis of 30 suspected meningitis patients were carried out using present available methods (Table 1) as well as PCR using 211 bp PorAgene amplicon asagenetic marker. The data shows false positive result with Gram (-ve) test (01 sample), glucose test (02 samples) and sucrose test (01 sample) while false negative result with oxidase test (02 samples) and catalase test (01 sample) which were confirmed by our method. For validation of the results, PCR was also carried out using two controls and bacterial culture of E. coli, Salmonella typhii andStreptococcus pyogenes (Table 2). The primers are specific and amplify only specific (211 bp) sequence of PorA and not other genes of different possible pathogens in the CSF. Therefore, specific 211 bp of PorAgene can be used as a genetic marker for quick diagnosis of bacterial meningitis.
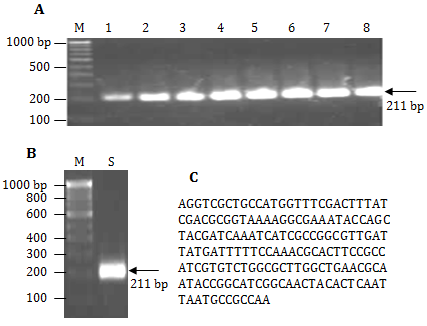
Figure 1 A. Agarose gel image of gradient PCR 100bp DNA ladder (lane M), PorA gene PCR amplicon (211 bp) at 51.0˚C, 52.2˚C, 54.4˚C, 57.3, 60.9˚C , 63.8˚C, 65.9˚C, and 67˚C (lane 1-8), B. 100bp DNA ladder (lane M), PorA gene amplicon (lane S), and C. Por A gen amplifies sequence (211 bp)
Results |
Microscopic |
Enzyme Test |
Latex Agglutination |
Biochemical Test |
PorA Marker |
||
Gram (-ve) |
Oxidase |
Catalase |
Glucose |
Sucrose |
|||
Positive |
4 |
2 |
1 |
3 |
5 |
4 |
3 |
Negative |
26 |
28 |
29 |
27 |
25 |
26 |
27 |
False Positive |
1 |
0 |
0 |
0 |
2 |
1 |
0 |
False Negative |
0 |
1 |
2 |
0 |
0 |
0 |
0 |
Table 1 Comparison analysis of thirty suspected patient samples of meningitis by diagnosis of using available methods and PorA marker
Samples |
Present Diagnosis Methods |
PorA |
Results |
|||||
Microscopic |
Enzyme Test |
Latex Agglutination |
Biochemical test |
|||||
Gram (-ve) |
Oxidase |
Catalase |
Glucose |
Sucrose |
||||
1 |
+ |
+ |
+ |
+ |
+ |
- |
+ |
C |
2 |
+ |
+ |
+ |
+ |
+ |
- |
+ |
C |
E. coli |
+ |
- |
+ |
- |
+ |
- |
- |
N |
S. typhi |
+ |
+ |
- |
- |
+ |
- |
- |
N |
S. pyogenes |
- |
- |
- |
+ |
+ |
+ |
- |
N |
Table 2 Cross reactivity test of PorA marker with two controls (containing Neisseria meningitidis) and three samples (containing Escherichia coli, Salmonella typhi, and Streptococcus pyogenes)
C: Control; N: Negative
The marker method can detect up to 2 ng of G-DNA or 9.36x104 CFU of N. meningitidis(1CFU~2.1x10-5 ng of G-DNA) directly from CSF of meningitis patients. This is very simple and economical method which can be used for quick detection of the bacterial meningitis caused by N. meningitidis from large number of patient samples at a time during an outbreak. The early diagnosis of the disease can save several lives. The 211 bp of PorAgene can be used in future for development of DNA sensor or microarray for the detection of bacterial meningitis due to the virulence nature of PorA gene.
None.
The author declares no conflict of interest.

©2017 Dash, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.