Journal of
eISSN: 2469 - 2786


Research Article Volume 8 Issue 2
1Department of R & D, Center for Genetic Engineering and Biotechnology, Cuba
2Department of Quality Control, Center for Genetic Engineering and Biotechnology, Cuba
Correspondence: José Miguel Fernández Torres, Department of R & D, Center for Genetic Engineering and Biotechnology, Cuba
Received: June 10, 2020 | Published: July 20, 2020
Citation: Torres JMF, Artiles YC, Ávila MTB, et al. Expression of the NS3 protein of hepatitis C virus used in the HCV UMELISA® test kit. J Bacteriol Mycol Open Access. 2020;8(2):55-59. DOI: 10.15406/jbmoa.2020.08.00273
To manufacturing the HCV UMELISA® Test kit, a recombinant NS3 protein is used as part of the antigen mixture. This protein is obtained from Escherichia coli XL-1Blue transformed with pR2M6-NS3-His plasmid downstream trp promoter. Regulation of expression has been identified as the main cause that affects yield and reproducibility at the fermentation process. To improve protein expression and transcription regulation, cloning of NS3 sequence was performed downstream tac promoter. At the same time, three E. coli strains (XL 1-Blue, W3110 and C600) were used to assess the expression of the NS3 protein. The tac promoter improved the expression level at all the strains evaluated from 20% under trp until 30 % and a reduction of promoter leakage before induction was observed compared with trp promoter. Due to the insensibility to the high metabolic burden, C600 was settled as the strain to use for NS3 expression. The new construction was well recognized by a pool of positive sera and individual sera from infected humans in an indirect ELISA using the HCV UMELISA® Test kit.
Keywords: HCV, C virus, ELISA, UMELISA, E. coli, hepatocellular, carcinoma
Hepatitis C is a disease that affects people causing liver cirrhosis and hepatocellular carcinoma. Its etiologic agent is an RNA virus named Hepatitis C virus (HCV).1 Screening for HCV infection is based on the detection of anti-HCV antibodies, and confirmed by the presence of HCV RNA to identify patients with ongoing infection.2 These tests are crucial to analyze the magnitude of the pandemic in different regions and design public health interventions. The NS3 protein of HCV is a multifunctional enzyme with serine-protease activity at the N-terminal end, and helicase activity at the rest of their structure that prevents coiling of RNA duplex strands formed during viral replication.3 In assays based on antibodies detection, the NS3 protein had been included due to its improvements in sensitivity and specificity on second generation tests.4 In Cuba, the ELISA for the screening of patients (HCV UMELISA® Test kit) is developed by the Center of Immunoassays. In this kit, an important biological reagent is a recombinant fragment of NS3 produced by the Center for Genetic Engineering and Biotechnology of Sancti-Spiritus. This protein is obtained from Escherichia coli XL-1Blue transformed with pR2M6-NS3-His plasmid downstream trp promoter on Luria-Bertani broth (LB). Regulation of expression has been identified as the main cause that affects yield and reproducibility at the fermentation process which leads to affect purification process. There are a few promoters that could be used to avoid leakage and to increase protein expression in complex media, like tac promoter.5 In this manuscript, we report the expression of this NS3 fragment downstream tac promoter to improve expression and promoter regulation in complex media.
Escherichia coli strains
XL1-blue: endA1 gyrA96(nalR) thi-1 recA1 relA1 lac glnV44 F'[:: Tn10 proAB+ lacIq (lacZ)M15] hsdR17(rK- mK+).
W3110: F- lambda- IN(rrnD-rrnE)1 rph-1
C600: F- tonA21 thi-1 thr-1 leuB6 lacY1 glnV44 rfbC1 fhuA1
Cloning the NS3-tac gene
The gene, encoding a stabilizing IL-2 fragment at the 5´ end linked to a fragment of HCV NS3 sequence, was extracted from pR2M6-NS3-His plasmid using the 5´ (aattccatggcgcctacttcaagttc) and 3´ (taagtcgacgcaggtgttgcaatcaat) oligonucleotides by PCR. The DNA fragment obtained was sub-cloned into the commercial vector pGEM-Teasy. Cloning of NS3- tac gene was released in Nco I and Sal I sites of PTBAD vector, which contains a polyhistidine tail at the Sal I site, digested previously with the respective enzymes (Nco I and Sal I) and dephosphorylated. The ligation reaction was done using T4 DNA ligase from pGEM-Teasy kit. So, the final construction would have incorporated a stabilizing IL-2 fragment of 60 amino acids in the protein and a polyhistidine tail for purification purposes. All cloning steps were carried out in Escherichia coli Xl-1 Blue. Grow kinetics of E. coli in 50mL shaking cultures and expression of recombinant protein Il2-NS3-His. The pR2M6-NS3-His and PTBAD-IL2-NS3-His vectors were used for the expression of IL2-NS3-His to evaluate expression of the protein under both trp and tac promoters respectively in three strains (XL-1Blue, W3110 and C600) on LB medium. PTBAD plasmid was used as negative control of expression. Inoculation in 5 ml LB medium supplemented with ampicillin (100µg/mL) was carried out with a single colony of the transformed strain with the corresponding plasmid and leaving it to grow at 37oC overnight. Then, 0.5ml of the culture was inoculated into 50mL of the LB medium supplemented with ampicillin and incubated at 37oC for 12 h while shaking at 250rpm. Induction of expression was done when cultures rises an optical density at 620nm (OD620nm) of 0.4-0.6, using IPTG (0.5mM) for PTBAD and PTBAD-IL2-NS3-His plasmids and β-Indoleacrilic acid (0.21mM) for pR2M6-NS3-His vector. Samples were collected periodically for evaluation.
SDS-page
Protein electrophoresis was performed on a 12% polyacrylamide denaturing gel with 2% Sodium Dodecyl Sulfate (SDS). Samples were diluted in loading buffer (125mM Tris-HCl, pH 6.8, 1% (w/v) SDS, 5% (v/v) glycerol, 10mM Dithiothreitol, 0.005% (w/v) Blue Bromophenol) and heated for 10min at 100 °C. The samples were run for about 45min at a constant voltage of 200V in running buffer (25mM Tris-HCl, pH 8.8; 192mM Glycine; 35mM SDS). The gel was stained with a 0.1% (w/v) Coomassie R-250 solution, 40% (v/v) methanol, 10% (v/v) acetic acid for 20min and the bands were visualized after gel destaining with 10% (v/v) methanol and 10% (v/v) acetic acid.
Detection of proteins by western blot
In Western blot assay, the proteins from the samples were transferred from the polyacrylamide gel to an extra Hybond-C nitrocellulose membrane (Amersham Life Science, UK) for immunodetection. As primary antibody, three monoclonal antibodies were used and dissolved at a concentration of 10μg/mL in a solution of skim milk (1% w/v) - PBS-Tween 20 (0.05% w/v). As the secondary antibody, an alkaline phosphatase conjugated mouse anti-IgG antibody (Sigma Aldrich, USA) diluted 1: 20 000 (Sigma Aldrich, USA) was used. Colorimetric detection was performed with 5-bromo-4-dichloro-indolyl phosphate / nitroblue tetrazolium (BCIP/NBT).
IMAC purification
From the total biomass obtained in a 500ml LB medium shaking flask of a growth culture, 0.6g were ruptured by sonication and centrifuged (10000rpm). Then the pellet was collected and solubilized (10ml/g biomass) for 1h in binding buffer pH 8 (8M urea, 100mM sodium phosphate) and filtered by 0.45µm. Once the matrix was equilibrated, the sample was passed through a "His-Select®" (GE Health care) matrix packaged in a C 10/10 column (GE Health care) with a height of 5cm. Elution was performed with the same buffer but at pH 6. The volumetric flow was kept constant at 0.6ml/min throughout the process. The chromatographic profile was monitored by measuring absorbance at 280nm. Denaturing electrophoresis in 12% polyacrylamide gel was used as a criterion of purity using image J V.0.5 program and protein concentrations was determined by BCA using bovine albumin as standard.6
ELISA for sensibility and specificity of recombinant NS3
To determine the sensitivity and specificity in the HCV UMELISA® Test kit we substituted the NS3 used in this kit by the purified NS3 expressed under tac promoter, in the mixture of antigens used in the coating solution. First was determined the optimum working concentration of the protein of interest that was of 1μg/mL and later were coated the plates using the mixture of antigens, and following all the recommended specifications to produce the diagnostic kit. For specificity, we used samples of negative sera from blood donors of the Havana Provincial Blood Bank. All samples were considered positive when the ratio (Fm-B)/ (P-B) was higher than or equal to 0.27. Where Fm, P and B refer to the fluorescence of the sample evaluated, the positive control concerning the HCV UMELISA® Test kit, and the blank, respectively. The sensitivity of antibody detection was estimated as the percentage of samples considered to be positive with the new NS3-tac protein concerning those considered to be positive with the NS3-trp referenced by a commercial kit (ORTHO® HCV 3.0). For this, 16 samples from seroconversion panels PHV 919 and 920 from SeraCare® were used. Specificity was defined as the percentage of negative HCV samples that were considered negative with the NS3-tac protein, compared to those considered negative with the NS3-trp protein in the HCV UMELISA® Test kit.
PCR products were analyzed on an agarose gel in parallel with Lambda Hind III-EcoRI DNA size marker and the expected 995bp fragment was observed (Figure 1A). Sequencing analysis was done to confirm amplification. At the PCR reaction, 12 amino acids were removed from the NS3 protein codified by pR2M6-NS3-His vector because it does not belong to viral NS3 sequence. The ligation reaction was done for cloning the amplified region into PTBAD vector. NS3 protein was expressed under both, trp promoter and tac promoter. A western-blot was performed for protein identification (Figure 1C). All fragments of the quimeric NS3 protein were recognized by the antibodies used on the assay. This showed that cloning NS3 under tac promoter was successful (Figure 1). A significant alteration on SDS-PAGE mobility between both proteins was observed. The NS3 expressed under tac promoter regulation migrated less than NS3 protein expressed downstream trp promoter although we expected the contrary. This anomalous migration pattern could be caused by altered SDS loading on each protein due to differences on the helicity at the C-terminal region among both proteins.7
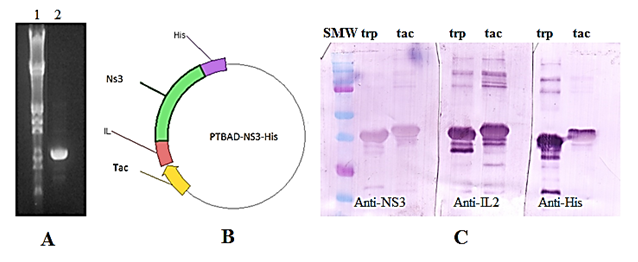
Figure 1 (A): PCR amplification of NS3 sequence. Line 1: lambda Hind III marker, line 2: PCR product (B): Scheme of the final vector for NS3 expression downstream tac promoter. (C): Western blot performed with NS3 transferred to a PVDF membrane and recognized by three monoclonal antibodies (The antibodies used are referred in the figure). SMW: Standard molecular weight. trp: quimeric NS3 protein expressed downstream trp promoter. tac: quimeric NS3 protein expressed downstream tac promoter.
NS3 expression
For evaluation of protein expression under both promoters, three strains were used as hosts and grown in 50ml shaking flasks (Figure 2). A reduction at growth rate was observed in the three strains (XL-1Blue, W3110 and C600) after promoter induction. The higher effect of protein expression on the final cell yield was observed on XL-1Blue strain. This is due to XL-1Blue has thi-1 mutation which impedes it to grow well without B1 vitamin. The same effect was seen on either W3110 and C600 strains but less than for XL. This growth decrease has been well described and is associated with the high metabolic burden when protein expression is achieved under strong promoters.8 The NS3 expressed under trp promoter in XL strain was 20% from total proteins like4 described. Protein expression under tac promoter was higher than 30% at each strain, whereas that downstream trp promoter was observed only an average of 20% in each strain. Other researchers have described too bigger expression with tac promoter than for trp promoter.5 Besides, the tac promoter allowed less protein production before induction than trp promoter in all strains assessed, showing better regulation on NS3 expression. The best regulation was observed in C600 strain when expressed NS3 downstream tac promoter with only 4.7% of protein leaking before induction while the worse regulation under tac promoter was observed on XL strain with 17.5%. This leakage on XL was not expected due to a lac Iq mutation present in their genotype. However, it is known that several amounts of lactose may be present in yeast extract and the tryptone.9 The lesser leaking of NS3 expression before induction in C600 strain is associated with a lacY- mutation in their genotype that prevent lactose permease synthesis and hence impede leakage corresponding to lactose induction. In terms of productivity, C600 showed major rate in NS3 production continued by W3110. The high expression, insensibility to big metabolic burden, great regulation of expression and major grow rate, settled C600 as the best strain for production of NS3 protein in LB medium downstream tac promoter.
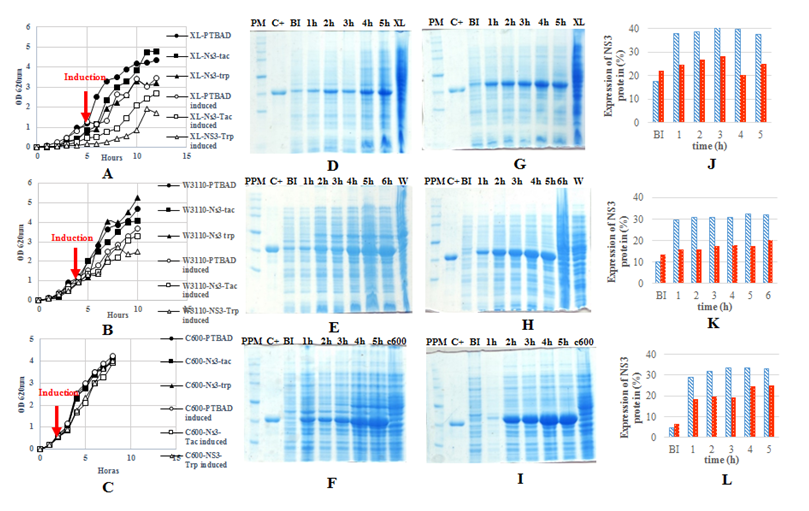
Figure 2 A & B & C: Growth kinetics in shaking flasks of the E. coli strains XL-1BLUE, W3110 and C 600 respectively. D & E & F: Polyacrylamide gel of the NS3 expressed downstream trp promoter in XL, W3110 and C600 strains respectively. G & H & I: Polyacrylamide gel of the NS3 expressed downstream tac promoter in XL, W3110 and C600 strains respectively (SMW: Broad range Protein marker, Promega, USA. C+: Purified NS3 used as a reference. BI: Sample was taken before induction. 1h,2h…5h: Hours after induction. XL: Xl-1Blue strain transformed with the plasmid without the insert. W: W3110 strain transformed with the plasmid without insert. C600: C600 strain transformed with the plasmid without the insert). J & K & L: Comparison of NS3 expression downstream both promoter (trp promoter in red and tac promoter in blue) in XL, W3110 and C600 respectively.
IMAC purification of NS3 protein
In diagnostic assays, the purity of antigens is crucial for avoiding cross-linking of antibodies from the sera. The new NS3 (NS3 Tac) was purified using metal affinity chromatography through the 6xHis tag attached to the C-terminus protein. Previously described in Palenzuela et al4 NS3 protein was expressed as a cytoplasmic inclusion body. Due to the insoluble nature of the expressed NS3 protein, urea was used as a denaturant. The protein eluted to pH 6 with a recovery of 59.9%. The purity obtained from the NS3-tac (97 %) is advantageous in the diagnostic test where the purity of antigens is required (Figure 3).
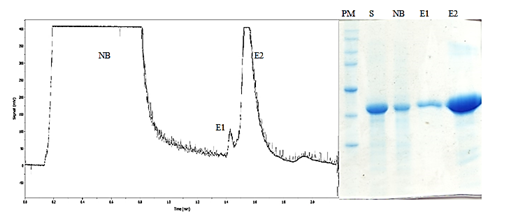
Figure 3 Chromatogram (A) and polyacrylamide gel of an IMAC purification of NS3 protein expressed downstream tac promoter in C600 strain. NB: Noun bound fraction. E1 and E2: Fractions eluted at pH6. PM: Broad range protein marker, Promega, USA. S: Sample before loading into the column.
Evaluation of the sensitivity and specificity of the recombinant protein
To assess the functionality of the protein, an HCV UMELISA® Test kit was used with the NS3 expressed under either tac or trp promoters coated both in a 96 well plate. For its recognition, the positive and negative sera supplied with the kit were used. As control was used a coated plate present in the kit, where the coating is performed with NS3 but also with other proteins of the nucleocapsid. Although a few amino acids were removed from the original sequence, was not observed differences between both proteins (Figure 4). Besides, a set of HCV positive samples and negative HCV samples from seroconversion panels PHV 919 and PHV 920 HCV SeraCare were used for its assessment. Here was used the purified NS3 protein (NS3 Tac) as a part of the antigen mixture in the coating solution and a reference mixture containing the NS3 obtained by the old strategy (NS3 downstream trp promoter). As control of specificity criteria, a commercial ORTHOÒ HCV 3.0 kit was used. The sensitivity of UMELISA with the new NS3 protein was 100 % in 16 samples of the PHV 919 and 920 HCV panels referenced by the ORTHOÒ HCV 3.0 kit (Table 1). These results show that the subtraction of the noun related amino acids on the NS3 protein does not affect the sensitivity and specificity in the HCV-UMELISA Test kit, therefore the new NS3 protein can be used in HCV diagnostic (Table 1).
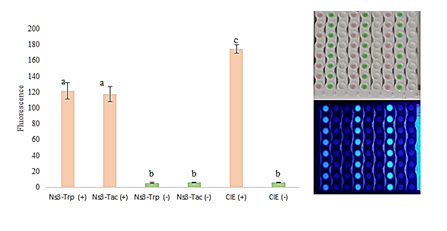
Figure 4 UMELISA performed with both NS3 expressed downstream both trp (NS3-Trp) and tac (NS3-Tac) promoters using the UMELISA HCV kit developed by Center of Immunoassays, Cuba. The negative and positive controls supplied with the kit were used and fluorescence values were compared by a test t (α<0,05)
Panel code |
NS3trp (Reference) |
|
NS3 tac(New) |
|
ORTHO HC:V 3.0 |
PHV 919-1 |
0_07 |
Neg |
0.06 |
Neg |
Neg |
PHV 919-2 |
0.09 |
Neg |
0.07 |
Neg |
Neg |
PHV 919-3 |
0.07 |
Neg |
0.06 |
Neg |
Neg |
PHV 919-4 |
0_10 |
Neg |
0.08 |
Neg |
Neg |
PHV 919-5 |
1.04 |
Pos |
0.94 |
Pos |
Pos |
PI1V 919-6 |
138 |
Pos |
1.26 |
Pos |
Pos |
PHV 919-7 |
126 |
Pos |
1.19 |
Pos |
Pos |
PHV 920-1 |
0_05 |
Neg |
0.04 |
Neg |
Neg |
PHV 920-2 |
0.11 |
Neg |
0.09 |
Neg |
Neg |
PHV 920-3 |
0_52 |
Pos |
0.59 |
Pos |
Neg |
PHV 920-4 |
1.04 |
Pos |
0.92 |
Pos |
Neg |
PHV 920-5 |
97 |
Pos |
0.93 |
Pos |
Pas |
PHV 920-6 |
12 |
Pos |
0.94 |
Pos |
Pos |
PHV 920-7 |
13 |
Pos |
139 |
Pos |
Pos |
PHV 920-1 |
134 |
Pos |
1.13 |
Pos |
Pos |
PHV 920-9 |
32 |
Pos |
33 |
Pos |
Pos |
Table 1 Comparison of serum diagnostic results obtained with the NS3 Trp and NS3 Tac proteins, showing the coating of HCV UMELISA Test kit and the certified sero conversion panels PHV 919 and 920 from Sera Care®
Relative fluorescence values were calculated as the (Fm-B)/(P-B) ratio(the samples are considered positives when this ratio is superior than 0.3), where Fm, P and B stand for the fluorescence of the samples, the positive control of the HCV UMELISA assay, and the blank, respectively. The presented values stand for the mean values of two replicates per serum tested
A new genetic construction was established, which enabled more than 30 % expression of the NS3 protein at the insoluble form in E. coli through genetic regulation via the tac promoter which avoided leaking of expression before induction comparing to trp promoter. Between the three strains assessed here, C600 was settled as the best host for NS3 expression downstream tac promoter. The new NS3 was successfully purified by IMAC and showed good activity in immunoassays when it was compared with the old NS3 used in HCV diagnostic by the HCV UMELISA® test kit.10
None.
The author declares that there is no conflict of interest.

©2020 Torres, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.