Journal of
eISSN: 2469 - 2786


Research Article Volume 12 Issue 2
1Department of Microbiology, Adekunle Ajasin University, Akungba-Akoko, Ondo State, Nigeria
2School of Engineering, University of Central Lancashire, United Kingdom
Correspondence: Dr. Oludare Temitope Osuntokun, Department of Microbiology, Adekunle Ajasin University, Akungba-Akoko, Ondo State, Nigeria
Received: June 13, 2024 | Published: July 5, 2024
Citation: Oludare TO, Stephen DO, Akele EO, et al. Emergence of pathogenic bacteria isolates from zea maize extract using 16s rRNA molecular sequencing protocol as a tools for microbial identification and characterization. J Bacteriol Mycol Open Access. 2024;12(2):50-57. DOI: 10.15406/jbmoa.2024.12.00373
The purpose of this research work is to determine the molecular identity of bacteria isolated from infected Zea maize using the 16s rRNA molecular sequencing protocol. The samples were obtained from Okeagbe, Akoko north-west local government in Ondo state with latitude and longitude of Okeagbe at 7.6450° N, and 5.7603° E respectively. Preparation of infected maize samples was cultured using the serial dilution method.. Confirmatory characterization of bacteria isolates using 16s rRNA (ribosomal RNA) sequencing procedures (purification, amplication, Sequencing, and DNA extraction) inclusive.The result shows the isolation of the bacteria isolates involved the culturing, inoculation, and plating of the isolate on a plated agar, the identification of the bacteria isolate includes the use of Gram staining, biochemical tests, and characterization using Bergey's manual and antibiotics Susceptibility Test. In Gram staining all bacteria isolates were positive except one, in the biochemical test most bacteria isolate was positive for sugar Fermentation and citrate test and all were negative for the Voges Proskauer test. In antibiotics Susceptibility test few were sensitive, most were susceptible to antibiotics used. With the use of the 16S rRNA and procedures (purification and application of product, Sequencing, and Extraction of DNA) the bacteria isolate were identified and characterized. The phylogenetic analysis and molecular identification of 16S rDNA sequencing revealed that Escherichia coli, Samonella enterica and Staphylococcus aureus were found to infect maize. Molecular characterization based on 16S rRNA Gene sequencing confirms the identity of bacteria. The conventional procedure shows the presence of different arrays of microorganisms in the infected maize, microbes identified are Bacillus subtilis, Bacillus anthracis, Micrococcus luteus, Clostridium sporogenes, Microbacterium lacticum, Clostridium sporogenes, Lactobacillus casei and Micrococcus luteus. The phylogenetic analysis and molecular identification of 16s rRNA sequencing revealed that Escherichia coli, Samonella enterica and Staphylococcus aureus were found to infect maize in Band fragment Base pair 1500bp. In conclusion, the hearsay that maize can only be infected by fungi, it was observed that the possibility of being infected with pathogenic bacteria is imminent. The bottom line is, there should be proper surveillance and food safety in our farm, market and food store, to prevent and totally eradicate emergence of pathogenic organism in our food item.
Keywords: 16s rRNA sequencing, molecular and conventional identification
Maize (Zea mays ssp. mays) originated in Mexico and Central America and belongs to the tribe Maydae of the family Poaceae. Maize from an old Greek name for a food grass. The term ‘maize’ seems to be derived from the word ‘mahiz’ in the Taino language of the Caribbean islands, which became ‘maiz’ in Spanish. The genus Zea consists of four species of which Zea mays L. is economically important. Maize is the world’s leading crop and is widely cultivated as cereal grain that was domesticated in Central America. It is one of the most versatile emerging crops having wider adaptability. Maize is one of the most vital food and industrial crops for human beings and is the most essential cereal crop across the globe after rice and wheat.1 In addition to its edible value, maize also serves as the raw material for industrial products and animal fodder.2,3 However, maize is susceptible to various pest diseases.4
Bacteria belonging to prokaryotes are ubiquitous and essential components of the Earth’s biota. Bacteria can be classified using conventional microbiology methods, such as microscopy, growth on specific media, biochemical and serological tests, and antibiotic sensitivity assays and also using Molecular methods such as 16S rRNA and polymerase chain reaction. Conventionally, culture-dependent microbiological methods were used to observe and enumerate viable microorganisms in a given sample. Molecular microbiology techniques have transformed bacterial identification in recent years.16s ribosomal RNA (rRNA) Gene sequencing is a common technique. In addition to being quicker and more accurate than conventional methods, this approach also enables the identification of bacterial strains to sub species level. The analysis of rRNA genes provides a framework for assigning sequences to genera and species, appropriate for investigating the microbial community's diversity.
Conventional detection of pathogenic bacteria is mainly based on cultivation procedures, which use enrichment broths followed by the isolation of colonies on selective media, biochemical identification, and confirmation of pathogenicity. This culture method is selective for the search for one type of pathogen at a time. Using the conventional method, Staley and Konopka in 1985 revealed only those bacteria, that are cultivable under lab conditions that exhibit only a smaller fraction of the microorganisms present in our environment are being studied and they called this phenomenon the “great plate count anomaly”. These findings proposed that microbiologists have succeeded only in discovering a small fraction of bacterial diversity from the huge microbial world, and many more things are yet to be discovered.4
When conventional methods were not reliable enough the advent of molecular biology in the 1980s contributed a set of powerful new tools that have helped microbiologists to detect the smallest variations within microbial species and even within individual strains. This has added an entirely new dimension to a science that was in danger of becoming constrained by its reliance on traditional laboratory techniques. Molecular techniques are major tools for the analysis of microorganisms from food and other biological substances. The techniques provide ways to screen for a broad range of agents in a single test.5 Molecular sequencing methods allow researchers to rapidly and specifically detect microorganisms of public health concern. Additionally, recent improvements have allowed simultaneous detection of several microorganisms in a single assay.6,7
Numerous discoveries and studies have demonstrated that fungi are the main pathogens responsible for the loss of economic importance of maize crops. It lowers the market value of the production of maize and results in an increase in the manufacturing of fungicides that have an antagonistic impact on fungi pathogens.7
Study designs and locations
Study designs and locations are (1) Okeagbe, Akoko north-west local government in Ondo state. The latitude and longitude of Okeagbe are 7.6450° N, and 5.7603° E respectively, (2) Supare Akoko, the southwest local government in Ondo state. The latitude and longitude of Supare Akoko are 7o27’8” N and 504’36” respectively. The samples obtained at the Okeagbe farm site were spoilt maize that has a feature of corn smut. The samples were obtained by chosen farm workers at the farm.
Sample collection
The first four samples (MS1- MS4) were collected at a farm site in Okeagbe Akoko, Akoko North-West Local Government, Ondo State, at approximately 5:25 p.m. 7.6450°N and 5.7603°E are the latitude and longitude, respectively. The other six samples (MS5 – MS 10) were taken at a farm site at 9:40 a.m. in Ondo State's Supare Akoko, Akoko Southwest local government. Supare is located at 7°27'8"N and 5°4'36E in latitude and longitude, respectively. Ondo State is located at latitudes of 5°45° and 7°52° and 4°20° and 6°05°, respectively. The sample was transported in a sterile Ziploc bag to the laboratory for further microbiological analysis.
Preparation of infected maize sample for culture protocol
9ml of distilled water was dispensed into 6 test tubes and the mouth was corked with cotton wool wrapped with aluminum foil and then sterilized at 1210C for 15 minutes using an autoclave. After sterilization, the water was allowed to cool for a few minutes, each test tube was then labeled as 10-1 - 10-6 respectively. 1gram of the infected maize sample was cut, weighed, and added to 9ml of sterile distilled water in a test tube and serially diluted by taking 1ml of the properly mixed solution with a sterile syringe and dispensed into the second diluent and this was done in an aliquot manner up to the fifth diluents.8
Identification of bacteria isolated from zea maize sample; macroscopic, microscopic examination, and biochemical characteristics of isolate
Preliminary identification and macroscopic analysis of the isolates were based on the cellular morphology characteristics which have a creamy pigmentation, irregular in shape, with a distinct colony, opaque. Cultural and microscopic examinations were done to identify the pure isolate. It also involves various and different biochemical tests which include: Gram staining, Simmons citrate, Lactose, Dextrose, Sucrose, Motility, Indole, Urease, Hydrogen Sulphide, Fructose, Maltose, Subitol, Mannitol, Glucose, Oxidase, Galactactase, Methyl red, V.P and Catalase tests were done for conventional identification of the isolates.9
Bacteriological analysis of the zea maize sample
The pour plate method of inoculation was used, and 1ml of the six-fold dilution of 10-3 and 10-5 samples (inoculums) was aliquoted into four sterile Petri dishes. Potato dextrose agar was prepared by dissolving 3.1 grams of the Agar, into 80ml of distilled water in a sterile conical flask, corked with cotton and aluminum foil, and then homogenized to dissolve. It was sterilized in an Autoclave at a temperature of 1210C for 15 minutes. After the sterilization, the medium was allowed to cool but not solidify. 20ml of Nutrient agar was then poured into different sterile Petri dishes containing the 1ml of the inoculums and allowed to set. Then the plates were incubated at 28oC for 72hrs.10 After 48 hours, the cultural characteristics on the plates were studied and recorded. The suspected colony was sub-cultured on fresh Potato dextrose agar and then incubated for 72 hours. Pure isolates were preserved on a double-strength potato dextrose agar slant for further studies.
Characterization of the isolate from zea maize sample
This entails comparing them to the results of the biochemical tests run on each of the bacterium isolates, which is done using Berger's manual as a preliminary procedure.11
Antibiotic susceptibility test of bacteria isolates from zea maize sample
The medium was then poured into appropriate Petri dishes aseptically. Antibiotic susceptibilities for Gram-positive bacteria and Gram-negative bacteria were determined according to the Clinical and Laboratory Standard Institute (CLSI) using the disc diffusion method. With the aid of an inoculating loop, suspected colonies were picked and inoculated in each plate and spread evenly to form a lawn culture, then the antibiotic discs were placed on the inoculated plates, and incubated at 37°C for 24 hours. The growth and zone of inhibition after 24 hours were recorded. The susceptibility test of the isolates was determined by using the following antibiotic discs; Pefloxacin (PEF, 10 µg), Gentamycin (CN, 10 µg), Ampiclox (APX, 30 µg), Zinnacef (Z, 20 µg), Amoxicillin (AM, 30 µg), Rocephin (R, 25 µg), Ciprofloxacin (CPX, 10 µg), Streptomycin (S, 30 µg), Septrin (SXT, 30 µg), Augmentin (AU, 30 µg), Tarivid (OFX, 10 µg). The antibiotic disc used was produced by Maxicare Medical Laboratory, Nigeria. The isolates were categorized as susceptible or resistant. The results were interpreted according to the Clinical Laboratory Standard Institute12 guidelines.
Molecular identification of the isolate using 16s rRNA gene sequencing protocol
Purification of amplified product of the bacteria isolates
After gel integrity, the amplified fragments were ethanol purified to remove the PCR reagents. Briefly, 7.6 µl of Na acetate 3M and 240 µl of 95% ethanol were added to each about 40µl PCR amplified products in a new sterile 1.5 µl tube Eppendorf, mixed thoroughly by vortexing and kept at 20°C for at least 30 min. Centrifugation for 10 min at 13000 g and 4°C followed by removal of supernatant (invert tube on trash once) after which the pellets were washed by adding 150 µl of 70% ethanol and mixing then centrifuge for 15 min at 7500 g and 4°C. Again remove all supernatant (invert tube on trash) and invert tube on paper tissue and let it dry in the fume hood at room temperature for 1015 min. then resuspended with 20 µl of sterile distilled water and kept at 20oC prior to sequencing. The purified fragment was checked on a 1.5% Agarose gel run on a voltage of 110V for about 1hr as previously, to confirm the presence of the purified product and quantified using a nano drop of model 2000 from Thermo Scientific.13
Molecular sequencing of the bacteria isolate
The amplified fragments were sequenced using a Genetic Analyzer 3130xl sequencer from Applied Biosystems using the manufacturers’ manual while the sequencing kit used was that of BigDye Terminator v3.1 cycle sequencing kit. Bio Edit software and MEGA 6 were used for all genetic analyses.14
Bacteria DNA extraction protocol
DNA was extracted using the protocol stated by Haque, et al.1 Briefly, Single colonies grown on medium were transferred to 1.5 ml of liquid medium, and cultures were grown on a shaker for 48 h at 28 ºC. After this period, cultures were centrifuged at 4600g for 5 min. The resulting pellets were resuspended in 520 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). Fifteen microliters of 20% SDS and 3 μl of Proteinase K (20 mg/ml) were then added. The mixture was incubated for 1 hour at 37 ºC, then 100 μl of 5 M NaCl and 80 μL of a 10% CTAB solution in 0.7 M NaCl were added and vortexed. The suspension was incubated for 10 min at 65 ºC and kept on ice for 15 min. An equal volume of chloroform: isoamyl alcohol (24:1) was added, followed by incubation on ice for 5 min and centrifugation at 7200g for 20 min. The aqueous phase was then transferred to a new tube isopropanol (1: 0.6) was added and DNA precipitated at –20 ºC for 16 h. DNA was collected by centrifugation at 13000g for 10 min, washed with 500 μl of 70% ethanol, air-dried at room temperature for approximately three hours, and finally dissolved in 50 μl of TE buffer.14
Molecular identification polymerase chain reaction
PCR sequencing preparation cocktail consisted of 10 µl of 5x GoTaq colorless reaction, 3 µl of 25 MgCl2, 1 µl of 10 mM of dNTPs mix, 1 µl of 10 pmol each 27F 5’ AGA GTT TGA TCM TGG CTC AG3’ and 1525R, 5′AAGGAGGTGATCCAGCC3′ primers and 0.3units of Taq DNA polymerase (Promega, USA) made up to 42 µl with sterile distilled water 8μl DNA template. PCR was carried out in a GeneAmp 9700 PCR System Thermal cycler (Applied Biosystem Inc., USA) with a Pcr profile consisting of an initial denaturation at 94°C for 5 min; followed by a 30 cycles consisting of 94°C for 30 s, 50°C for 60s and 72°C for 1 minute 30 seconds; and a final termination at 72°C for 10 mins. And chill at 4oC.GEL. Gel electrophoresis, PCR amplicon purification, sequencing, and editing are as previously.13
The samples obtained in total were eight samples and a total number of ten bacteria isolates were isolated from the infected maize samples.
Table 1: It shows the macroscopic morphological characteristics of the organism isolated from the Zea maize sample collected, which includes the shape, the colour, growth diameter, and shape.
|
Isolate code |
Shape |
Surface pigmentation |
Bottom pigmentation |
Edge |
|
MS1 |
Rod |
Cream |
Cream |
Irregular |
|
MS2 |
Irregular |
Cream |
Cream |
Irregular |
|
MS3 |
Irregular |
Cream |
Cream |
Irregular |
|
MS4 |
Rod |
Cream |
Cream |
Irregular |
|
MS5 |
Rod |
Cream |
Cream |
Irregular |
|
MS6 |
Rod |
Cream |
Brown |
Curved |
|
MS7 |
Rod |
Cream |
Cream |
Irregular |
|
MS8 |
Irregular |
Cream |
Brown |
Curved |
|
MS9 |
Rod |
Cream |
Cream |
Irregular |
|
MS10 |
Irregular |
Cream |
Cream |
Irregular |
Table 1 Morphological characteristic of the bacteria isolate from zea maize sample
Key: MS (Maize Sample)
Table 2: It shows a preliminary test carried out on the spoilt maize sample isolate. The test includes Motility, Sugar fermentation, Indole, Lactose, Dextrose, and Sucrose. Urease, Hydrogen sulphide, Gas production. It was observed that all five isolates were positive for Simmon citrate, All isolate was positive for dextrose, all isolate was positive for catalase, all isolate were also positive for oxidase, all isolate was positive for maltose, all isolate was positive for fructose. All isolates were negative methyl red, urease, motility, sucrose, and also for gas production. All isolate were negative except for MS3 which was positive for lactose, while all was positive except for MS2 which was negative for glucose. All isolate was positive except for MS1 was negative for sorbitol, Also MS4 was the only negative out of all isolate for mannitol and MS5 was the only isolate negative for V.P while other isolate was positive.
|
Isolate code |
Motility |
Indole |
Urease |
H2S |
Gas |
Lactose |
Dextrose |
Sucrose |
Citrate |
Fructose |
Maltose |
Subitol |
Mannitol |
Glucose |
Oxidase |
Galactose |
Metyl red |
Vp |
Catalase |
|
MS1 |
+ |
- |
+ |
- |
+ |
- |
+ |
+ |
+ |
+ |
+ |
- |
+ |
+ |
+ |
+ |
- |
+ |
+ |
|
MS2 |
+ |
+ |
+ |
- |
+ |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
- |
+ |
= |
- |
+ |
+ |
|
MS3 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
- |
+ |
+ |
|
MS4 |
+ |
- |
+ |
- |
+ |
- |
+ |
+ |
+ |
+ |
+ |
+ |
- |
+ |
+ |
+ |
- |
+ |
+ |
|
MS5 |
+ |
+ |
+ |
- |
+ |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
- |
- |
+ |
|
MS6 |
+ |
+ |
+ |
- |
- |
+ |
+ |
+ |
+ |
- |
+ |
_ |
+ |
+ |
+ |
- |
- |
- |
+ |
|
MS7 |
+ |
- |
+ |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
- |
+ |
+ |
|
MS8 |
+ |
+ |
+ |
+ |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
- |
+ |
+ |
+ |
- |
+ |
+ |
|
MS9 |
+ |
- |
+ |
+ |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
- |
+ |
+ |
|
MS10 |
+ |
+ |
+ |
+ |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
- |
+ |
- |
Table 2 Shows the biochemical characteristics of the bacteria isolate from zea maize sample
Key: + (Positive), - (Negative), MS (Maize Sample).
Table 3: Shows the result of Gram staining and the microscopic examination of the isolates. It was observed in this table that the Maize sample 1, maizes sample2, Maize sample 3, maize sample 4, maize sample 5, maize sample 6, maize sample 7, and maize sample are all positive to Gram staining while maize sample 8 and maize sample 9 are negative to Gram staining reaction.
|
Isolates code |
Gram stain |
Color |
|
MS1 |
+ |
Purple |
|
MS2 |
+ |
Purple |
|
MS3 |
+ |
Purple |
|
MS4 |
+ |
Purple |
|
MS5 |
+ |
Purple |
|
MS6 |
+ |
Purple |
|
MS7 |
+ |
Purple |
|
MS8 |
- |
Pink |
|
MS9 |
- |
Pink |
|
MS10 |
+ |
Purple |
Table 3 Gram stain and microscopic examination on the isolates
Key: + (Positive), - (Negative), MS ( Maize Sample).
Table 4: The probable organisms include Bacillus subtilis, Micrococcus luteus, Clostridium sporogenes, Macrobacterium lacticum, Lactobacillus casie, Bacillus anthracis2
|
Isolated code |
Probable organism |
|
MS1 |
Bacillus subtilis |
|
MS2 |
Micrococcus luteus |
|
MS3 |
Bacillus subtilis |
|
MS4 |
Clostridium sporogenes |
|
MS5 |
Microbacterium lacticum |
|
MS6 |
Bacillus anthracis |
|
MS7 |
Clostridium sporogenes |
|
MS8 |
Lactobacillus casei |
|
MS9 |
Bacillus anthracis |
|
MS10 |
Micrococcus luteus |
Table 4 Characterization of the bacteria isolate from Zea maize sample
Key: MS (Maize Sample).)
Figure 1: Shows the antibiotic susceptibility test of the isolate after being Gram stained. It was observed that Bacillus subtilis has the highest zone of inhibition of (20mm) in ciproflox and the lowest zone of inhibition of (10mm) in streptomycin, Micrococcus luteus has the highest zone of inhibition of (20mm) in pefloxacin, and the lowest zone of inhibition of (10mm) in ciproflox. Bacillus subtilis has the highest zone of inhibition of (20mm)in rocephin and has lowest zone of inhibition of (10mm) in ampliclox, while Clostridium sporogenes have the highest zone of inhibition of (15mm) in gentamycin and its lowest zone of inhibiting is (12mm) in ampliclox. Macrobacterium lacticum has the highest zone of inhibition to ciproflox (18mm) while is lowest zone of inhibition to amoxicillin (9mm). Clostridium sporogenes has the highest zone of inhibition (17mm) in Ampliclox and the lowest zone of inhibition (7mm) in Gentamycin. Lactobacillus casei has the highest zone of inhibition (15mm) in Erythromycin and the lowest zone of inhibition (10mm) in rocephin. Micrococcus luteus has the highest zone of inhibition (18mm) in pefloxacin and the lowest zone of inhibition (10mm) in Ampliclox. Bacillus subtilis, Micrococcus luteus and bacillus anthracis has no zone of inhibition in Ampliclox, bacillus subtilis has no zone of inhibition in both amoxicillin and ampliclox. Also, Macrobacterium lacticum has no zone of inhibition in pefloxacin, erythromycin, and streptomycin.
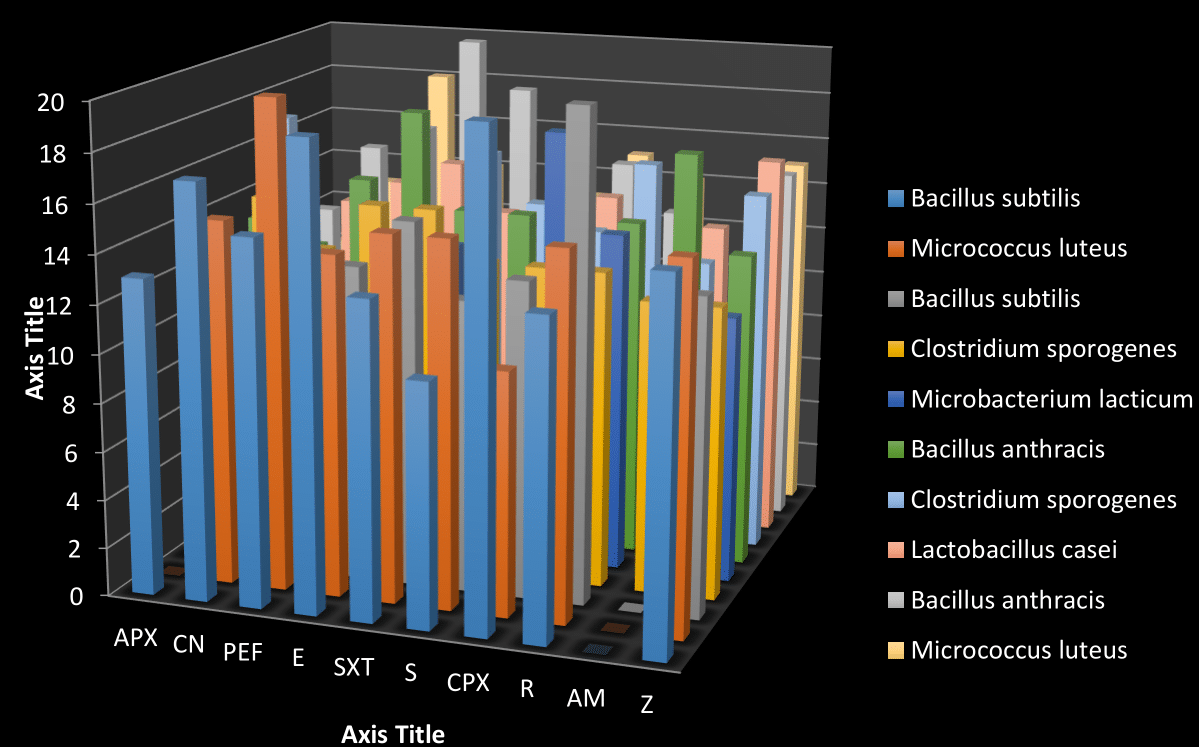
Figure 1 Antibiotic susceptibility of bacteria isolate from zea maize sample.
KEY: S, Streptomycin; E, Erythromycin; PEF, Pefloxacin; CN, Gentamycin; CPX, Ciproflox; SXT, Septrin; AM, Amoxicillin; R, Rocephin; Z, Zinnacef; Apx, Ampliclox.
UNIT: Milimetre (mm).
Molecular Identification of Bacteria Isolate Using 16S rRNA Sequencing Protocol
|
Sample ID |
Scientific Name |
Max score |
Total score |
Query cover |
E value |
Per. ident |
Accession |
|
E1 |
Escherichia coli |
2656 |
2656 |
99% |
0 |
100.00% |
|
|
E2 |
Salmonella enterica subsp. Arizonae |
2660 |
2660 |
100% |
0 |
100.00% |
|
|
E3 |
Staphylococcus aureus |
2702 |
2702 |
99% |
0 |
99.86% |
Table 5 Identified bacteria isolate using 16S rRNA gene sequencing protocol
The purpose of this research work is to determine the molecular identity of bacteria isolated from infected Zea maize using the 16s rRNA molecular sequencing protocol. The research has demonstrated that there is no antagonistic relationship between bacteria and fungi when they coexist.15,16 The infected maize was analyzed using the conventional method (Gram staining and biochemical test) and molecular method (16S rRNA Gene sequencing). It is known that the successful cultivation of bacteria is highly dependent on the distribution in the environment and on the growth state of the bacteria at the time of sampling, whether they are active or starved.17
The bacteria isolates from infected maize were Gram-positive, the biochemical test signifies negative to Voges-Proskauer test and positive to methyl red test. This might be due to the inability of organisms to produce the acetylmethylcarbinol (acetoin) on the digestion of glucose. They were also positive for sugar tests like sucrose, fructose, dextrose, glucose, and lactose while few were negative for to indole test which is a variation in some biochemical parameters. These findings are also similar to those reported by Moreno, et al.18
Bacterial phenotypic characterization using the biochemical identification of the isolates included in this study showed that some biochemical differences exist among the different species but in some cases, they were not sufficient to allow for a differential identification. This is a known drawback to the identification of bacteria isolates using conventional methods. These particular characteristics are intra-species variability, inter-species similarity, and bias toward previously recognized bacterial species. Antibiotic susceptibility tests showed low sensitivity for Ampicillin and least sensitivity for Streptomycin. Further, the tested organism also showed sensitivity to Erythromycin and Gentamycin. These results are found identical reported by Moreno, et al.18
For effective pathogen identification, many detection and typing techniques have been created and are now extensively used. The discriminatory power of the utilized detection or typing technique is significantly increased by the combination of two or more primers and/or methods and by method optimization. Because of their increased specificity, sensitivity, simplicity of use, and speed of detection, molecular technique advancements offer a competitive advantage over other approaches currently used to identify bacteria in samples. While molecular techniques and their more sophisticated equivalents can produce findings in as little as a few hours, the majority of culture-independent methods rely on the study of microbial nucleic acid sequences (DNA and/or RNA). The majority of them rely on the 16S rRNA gene sequence and polymerase chain reaction (PCR) method to amplify these nucleic acids. The bulk of microorganism diagnosis amplification methods rely on the rRNA genes. (rDNA).18 In the context of rRNA target sequences, It has truly come of age and its range of application is perceived to broaden shortly. The food industries, water processors, and analytical laboratories have taken up the latter method; for rapid differentiation of species, strain identification, and definition of strain relatedness from infected samples.5
In this study, prompt and accurate identification of microorganisms, using 16s rRNA plays a vital role in the identification of the infected Zea maize. It was observed that different organisms were identified and named with the molecular method. Bacillus subtilis, Bacillus anthracis, Micrococcus luteus, Clostridium sporogenes, Microbacterium lacticum, Clostridium sporogenes, Lactobacillus casei and Micrococcus luteus were identified using 16s rRNA molecular sequencing this organisms are rather very pathogenic in nature.
Most medical microbiologists rely on conventional techniques (microscopic examination, for example, the Gram staining test, culture on selective media, and biochemical tests, for example, catalase test, and indole test) to identify microorganisms.19 Often, identification using conventional techniques is not very accurate, leading to misidentification of the Microorganisms.19 For well-established, well-known bacterial species, phenotypic identification is known to be more reliable; nevertheless, species-level identification might be challenging for uncommon or less well-known species. With the use of 16S rRNA gene sequencing, this restriction can be overcome to a large extent. Generally, sensitive and rapid detection of bacteria in samples is now possible in a largely positive way because of the major advances, use of 16s rRNA Gene sequencing.20–22 Evaluated the suitability of 16S rRNA for the identification of bacteria strains of Gram-positive rods.
The phylogenetic analysis and molecular identification of 16S rDNA sequencing revealed that the organisms Escherichia coli, Salmonella enterica and Staphylococcus aureus were found to infect maize. This study has shown that Escherichia coli, Salmonella enterica, and Staphylococcus aureus are potential bacteria that cause in maize crops infestation.
This study indicates the utility and value of the 16S rRNA gene sequence as a tool for quick and precise initial screening of bacteria in Zea maize samples and for reliable assessment of phylogenetic relatedness. The finest understanding of the variety, structure, and function of the microbial communities were done by 16S rRNA Gene sequencing. In this study, it has also shown that bacteria infect maize. The characterization of the bacteria isolated from maize was performed using various morphological, biochemical, and physiological parameters. Molecular characterization based on 16s rRNA Gene sequencing confirms the identity of bacteria. The results indicate the identity of the bacteria that infect Maize crops. In conclusion, the assumption that only fungi is found in Zea maize is almost a myth, this research validate that pathogenic organisms may also be found in infected Zea maize, imminent. The bottom line is, there should be proper surveillance and food safety in our farm, market and food store, to prevent and totally eradicate emergence of pathogenic organism in our food item.
More research studies should be carried out on maize crop pathogens. The use of 16S rRNA gene sequencing should be frequently used in research studies, so as to get more reliable and accurate results during the identification of bacteria.
Self-funded research, authorship, and/or publication of this article: Financial assistance was not provided by any Research Foundation.
The laboratory staff of Adekunle Ajasin University, Department of Microbiology, Faculty of Science, Akungba Akoko, Ondo State, Nigeria.
The authors declare that there are no conflicts of interest.

©2024 Oludare, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.