Journal of
eISSN: 2469 - 2786


Research Article Volume 4 Issue 4
1Grupo de Investigacion en Acuicultura (GIA), Universidad de Las Palmas de Gran Canaria, Spain
2Servicio de Microbiologia, Instituto de Investigacion Marques de Valdecilla IDIVAL, Spain
3Instituto Universitario de Sanidad Animal, Spain
Correspondence: Dipanshu Kumar, Department of Paedodontics and Preventive Dentistry, Institute of Dental studies and Technologies, Kadrabad, Modinagar, 66c, Pocket-A, Mayur Vihar, Phase II, New Delhi, India, Tel 9013190478
Received: April 12, 2017 | Published: April 28, 2017
Citation: Belinda V, José RV, Jimena B, et al. Effect of different culture conditions on the (twitching) displacement of Photobacterium damselae subsp. piscicida. J Bacteriol Mycol Open Access. 2017;4(4):134-141. DOI: 10.15406/jbmoa.2017.04.00103
Photobacterium damselae subsp. piscicida (Phdp), etiological agent of photobacteriosis, is a non-motile bacterium. Notwithstanding, it is able to move on solid surfaces. The presence of pili-like structures on the bacterial surface has been recently described and being associated with twitching displacement by our team. The objective of this study was to modify some conditions (temperature, pH, concentration of nutrients, salinity, solidification of the medium, surface roughness and presence of target cells of Phdp) to determine which ones promote motility on solid surfaces. Lower temperatures (≤18°C), alkaline and acidic environments and lacking salinity (<0.5%) media had a negative effect on cell motility, while nutrient limitation did not affect the bacterial response. The best twitching motility was observed when Phdp was inoculated in medium with 0.2% agar and pH 7.0. Also, interesting results were obtained on rough surfaces or by the addition of cellular debris of SAF-1 cell line to the medium.
According to the results, the “new” composition of the “twitching displacement” medium could be as follows: 3.7% (w/v) brain-heart infusion broth (BHIB) supplemented with about 1.0% (w/v) NaCl and 0.2% (w/v) agar, with a final pH of 7.0, and maybe, scrape petri dishes and add debris of SAF-1 cells 1:2-factor dilution to medium. If bacterial strains are incubated at 22-25°C for three days, a good response should be achieved in terms of fimbriae production.
Keywords: photobacterium damselae subsp. Piscicida,twitching, bacterial displacement, culture media
Phdp, photobacterium damselae subsp. piscicida; ATCC, american type culture collection; spp., species; Subsp., subspecies; PBS, phosphate buffered saline solution; BHIB, brain-heart infusion broth; BHIA, brain-heart infusion agar; SAF-1, cell line from sparus aurata fin; NaCl, sodium chloride; CV, crystal violet; PCR, polymerase chain reaction; RT, room temperature; OD600, optical density at 600nm; p, p-value; w/v, weight/volume; ml, milliliter; nm, nanometre; h, hour; °C, degree celsius; %, percent
Photobacterium damselae subsp. piscicida (Phdp), which belongs to the family Vibrionaceae, is a pleomorphic Gram-negative bacterium, with bipolar staining, non-motile and halophilic.1,2 It grows at 15-30˚C, but the disease caused by this organism usually occurs when water temperatures are over 20˚C.
The etiological agent of fish photobacteriosis or pasteurellosis3 is an important pathogen affecting different fish species in Europe, Japan, USA and Mediterranean countries.4,5 It first appeared in USA in 1963 affecting wild white perch (Morone americanus) and striped bass (Morone saxatilis).6 Afterwards, it caused serious economic losses in aquaculture in Japan7 and from the early 1990s began to spread across Europe, becoming a major problem in the global marine aquaculture.
Different studies reveal that Phdphas virulence factors involved in its pathogenesis: presence of a polysaccharide capsular layer8,9 plasmid-encoded virulence factors,10 iron uptake mechanisms,11 proteins with apoptotic activity,12 other enzyme activities,13,14 and resistance to blood serum of several fish species.15
“Twitching motility”, term introduced is a kind of motion mediated by a proteinaceous and filamentous structure called Type-IV fimbriae or pili.16,17 It is located on the surface of many Gram-negative and Gram-positive bacteria, at one or both poles.18-20 And also, it is involved in surface adhesion and colonization,21-23 tropism determination,24 biofilm formation,23 genetic material uptake21 and virulence of bacteria.25-27 Fimbriae expression is controlled by transcriptional and post-transcriptional28 and is influenced by several factors: bacterial growth rate,29 culture medium composition,30,31 temperature,32 pH and osmolarity.33 So, it is a complex and unstable process and its levels are heterogeneous, frequently becoming lost.34
Recently, the presence of surface appendages similar to pili on Phdp (Figure 1) and the ability of this pathogen to move on solid surfaces have been described by our team.35 Thus, we aim to go into detail about this subject, starting by studying and determining what factors and environmental conditions affect twitching displacement of Photobacterium damselae subsp. piscicida. Identifying these parameters should contribute to a better understanding of the correlation between the adherence capabilities and the pathogenicity of this bacterium.
Ethical approval is not required by a specific committee since animals has not been used in the present study for research purposes.
Bacterial strains
The five Photobacterium damselae subsp. piscicida strains used in this study are listed in Table 1. These bacteria were verified by biochemical and PCR tests and preserved in brain-heart infusion broth (BHIB-1.5) with 1.5%NaCl and 25% glycerol at -80˚C until use.
Phdp C2 |
Gilthead seabream (Sparus aurata) |
Spain, 1997 |
|
Phdp 94/99 |
Gilthead seabream (Sparus aurata) |
Spain, 1999 |
|
PhdpDI21 |
Gilthead seabream (Sparus aurata) |
Spain, 1991 |
|
PhdpPP3 |
Yellowtail (Seriolaquinqueradiata) |
Japan |
|
Phdp ATCC 17911 |
White perch (Roccus americanus) |
USA, 1964 |
Table 1 Bacterial strains used in this study
Prior to experiment
The strains were routinely cultured in brain-heart infusion agar (BHIA-1.5) supplemented with 1.5% NaCl and incubated at 22˚C for 24 hours. For experiments, 1-5 colonies of each strain were passed to 10ml of brain-heart infusion broth (BHIB-1.5) supplemented with 1.5% NaCl and incubated at room temperature (RT), in darkness for 24h. When the medium was turbid (OD600≈0.45), it was diluted 1:100 with sterile phosphate buffered saline (PBS) solution, becoming the inoculum of the experiments.
“Twitching displacement” medium
The composition of the medium used to the experiments was brain-heart infusion broth supplemented with 1.5% NaCl and 0.3% agarose D-1 Low EEO (CONDA, Pronadisa). It was sterilized at 120˚C for 20 minutes, prepared and poured into Petri dishes the day before the experiment and kept in darkness at RT until use.
Displacement assays
After preparing everything, we proceeded to the inoculation of the media. To do this, sterile flat toothpicks were immersed into the bacterial suspensions for about 5 seconds and inoculated in three points into the agar to touch the plate underneath the agar, being removed after 1-2 seconds. Afterwards, it was incubated for 48 and 72 hours in darkness.
The length of bacterial shifts from inoculation point was measured after 2 and 3 days of incubation in every experiment to determine what environmental conditions favor “twitching motility”. These conditions are detailed below.
Temperature of incubation
The temperatures of incubation studied were 18, 20, 22 (reference temperature) and 25˚C. And furthermore, some agar plates were kept at 4˚C for 3 and 7 days and later, inoculated and incubated at 22˚C for other 2 and 3 days.
pH
The culture medium was prepared in the same conditions described above and the pH was adjusted with phosphate buffer to a final pH of 6, 7, 8 and 9.
Concentration of nutrients
The quantity of brain-heart infusion broth was reduced by half and a quarter to evaluate the bacterial response when the availability of nutrients decreases.
Salinity of the medium
For this assay, the medium was supplemented with 0, 0.5, 1, 1.5, 2 and 2.5% of sodium chloride (NaCl). Also, we inoculated in “salinity medium” with 0% NaCl (0.3% agarose, 0.1% yeast extract, 0.4% peptone; w/v).
Agar concentration of the medium
0.3% agar added to the medium was compared to 0.2 and 0.4% to check whether the degree of solidification of the medium facilitates or curbs bacterial motility.
Scraping Petri dishes
The objective of this test was to observe whether rough surfaces benefit the biofilm formation and cell attachment, as well as compare the motility on rough and smooth surfaces. It was carried out under sterile conditions before serving the “motility” medium by using sterile scissors to draw a “W” on the plate in every direction.
Cell debris in the medium
Debris of SAF-1 cells was added in the medium in order to generate tropism, previously knowing that Phdp is able to adhere to, invade and survive within these cells.36 This cell line consists of fibroblastic cells from fin of gilthead seabream (Sparus aurata).37 Cells from culture bottles were diluted with PBS to 0.409 optical density at 600 nm. Next, it was diluted by a factor of 1:2, 1:4 and 1:8. 10 ml of each dilution were mixed with 200 ml of medium.
Crystal violet staining
After three days of incubation, agar was carefully removed from petri dishes to show the bacterial growth and shift on the plate beneath the agar. Loose cells and agar remains were washed with distilled water and then, the attached bacteria were stained with crystal violet (CV) solution (0.7% w/v). Twelve minutes later, it was washed and air dried. Finally, we could observe the bacteria fixed to the plate.
Statistical analysis
Analyses were produced on IBM SPSS Statistics 22.0 (IBM Corp.), using general linear models: multivariate (posthoc tests: Tukey, Duncan and Hochberg) to compare the “different groups” of each assay for both data collection days (days 2 and 3 of incubation) separately and univariate to analyse the differences on motility between “days 2 and 3 of incubation” for each group. Models indicate significance when p≤0.05.
Temperature of incubation
Strains C2 and ATCC 17911 developed displacement at all temperatures tested, with no significant difference (p˃0.05). The same for strains PP3 and DI21when Phdp was incubated for 3 days, but on the second day of incubation, we could see no growth and no motility at 18˚C (p˃0.05). Nevertheless, strain 94/99 had not motion at 18˚C (day 2) nor when it was previously kept cold for a week (days 2 and 3) (p˃0.05) (Figure 2). There were no significant differences between days of incubation either (p˃0.05), except for groups “7 days at 4˚C” (strain DI21) (p˂0.01) and “18˚C” (strain ATCC 17911) (p˂0.04) (Figure 2).
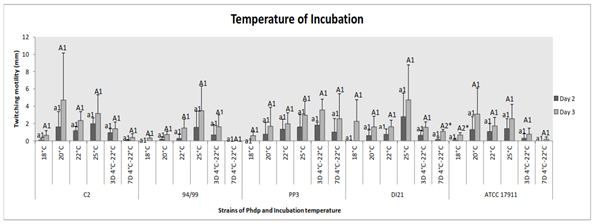
Figure 2 Effect of temperature of incubation on twitching motility of the Phdp strains tested: C2, 94/99, PP3, DI21 and ATCC 17911.
Small letters: Compare “groups” after 48 h of incubation
Capital letters: Compare “groups” after 72 h of incubation
Numbers: Compare “days of incubation” of each group
*Indicates that there are statistically significant differences
pH
After 2 days of incubation, all the strains only swelled and moved at pH 7, with significant difference in the case of C2 (p˂0.005) and DI21 strains (p˂0.05). After 3 days, history repeated itself for strains C2, 94/99 and ATCC 17911, with significant difference for the first one (p˂0.0001); while the other strains had displacement at different pH, although with significant difference between pH 7 and other pH values (strain PP3) (p˂0.005) (Figure 3). Comparing days of incubation, there were significant differences for groups “pH 7” (strain C2) (p˂0.03) and “pH 6” (strain DI21) (p˂0.04) (Figure 3).
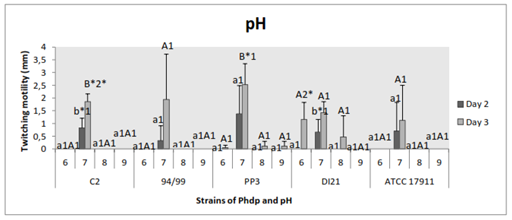
Figure 3 Effect of pH on twitching motility of the Phdp strains tested: C2, 94/99, PP3, DI21 and ATCC 17911.
Small letters: Compare “groups” after 48 h of incubation.
Capital letters: Compare “groups” after 72 h of incubation.
Numbers: Compare “days of incubation” of each group.
*Indicates that there are statistically significant differences.
Concentration of nutrients
Bacteria showed displacement at all concentrations of nutrients tested, with no statistically significant differences between groups (p˃0.05) and between days of incubation (p˃0.05). These results were obtained in all cases (Figure 4).
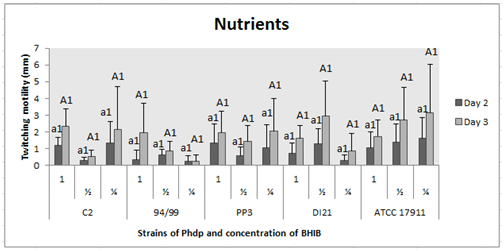
Figure 4 Effect of nutrients on twitching motility of the Phdp strains tested: C2, 94/99, PP3, DI21 and ATCC 17911.
Small letters: Compare “groups” after 48 h of incubation.
Capital letters: Compare “groups” after 72 h of incubation.
Numbers: Compare “days of incubation” of each group.
*Indicates that there are statistically significant differences.
Salinity of the medium
Regarding strain C2, we found significant difference on displacement between bacteria inoculated in medium supplemented with 1% NaCl and media containing no added NaCl (p˂0.01). Between groups “NaCl free-salinity medium”, wherein bacteria had no growth or motility, and “twitching motility medium with 0% NaCl”, between every group and between days of incubation, there were no significant differences (p˃0.05) (Figure 5). The following strain, 94/99, had no significant difference between groups (p˃0.05) and between days of incubation (p˃0.05), just the same as strains PP3, DI21 (except between days 2 and 3 of incubation in Group “1%” (p≤0.05)) and ATCC 17911 (except between days in Group “2.5%” (p˂0.01)) (Figure 5).
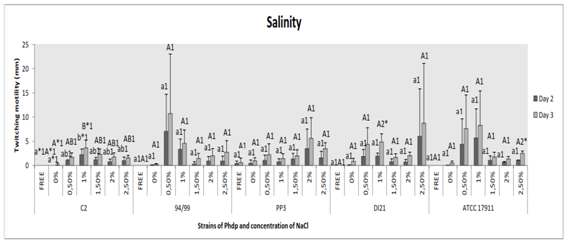
Figure 5 Effect of salinity of the medium on twitching motility of the Phdp strains tested: C2, 94/99, PP3, DI21 and ATCC 17911.
Small letters: Compare “groups” after 48 h of incubation
Capital letters: Compare “groups” after 72 h of incubation
Numbers: Compare “days of incubation” of each group
*Indicates that there are statistically significant differences
Agar concentration of the medium
Significant increased motility was observed when the concentration of agar was lower (0.2% w/v) in strains 94/99 (p˂0.02), DI21 (p≤0.05) and ATCC 17911 (p≤0.05); the other two strains also moved larger in 0.2% agar-medium, but with no statistical significance (p˃0.05) (Figure 6). Significant difference between days of incubation was not detected (p˃0.05) (Figure 6).
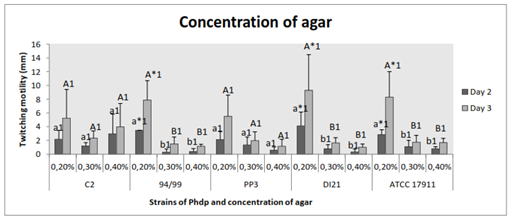
Figure 6 Effect of agar concentration of the medium on twitching motility of the Phdp strains tested: C2, 94/99, PP3, DI21 and ATCC 17911.
Small letters: Compare “groups” after 48 h of incubation.
Capital letters: Compare “groups” after 72 h of incubation.
Numbers: Compare “days of incubation” of each group.
*Indicates that there are statistically significant differences.
Scraping petri dishes
Scrape the petri dishes or not was not statistically different (p˃0.05). However, displacement was slightly higher for all strains in the first case. We could always notice that the bacterial shifts were larger on the third day of incubation as in all assays, except when there was no motility; at this time, with no significant difference (p˃0.05) (Figure 7).
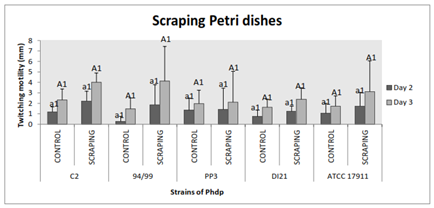
Figure 7 Effect of scraping the area of bacterial shift and attachment on twitching motility of the Phdp strains tested: C2, 94/99, PP3, DI21 and ATCC 17911
Small letters: Compare “groups” after 48 h of incubation
Capital letters: Compare “groups” after 72 h of incubation
Numbers: Compare “days of incubation” of each group
*Indicates that there are statistically significant differences
Cell debris in the medium
There were no significant differences between the groups “control” and “presence of cells in the medium” (p˃0.05) and between the different dilutions of cells (p˃0.05), for any strain. In the case of strains 94/99, DI21 and ATCC 17911, displacement best values seemed to be associated with presence of cell but in a lesser proportion, whereas for strain PP3 it was the opposite, displacement was not observed at 1:8 factor- dilutions. Strain C2 had groups with similar results (p˃0.05) (Figure 8). Significant differences between days of incubation in groups “control” (94/99 and DI21) (p≤0.05) and “1:4 factor-dilution” (DI21) (p˂0.01) were obtained (Figure 8).
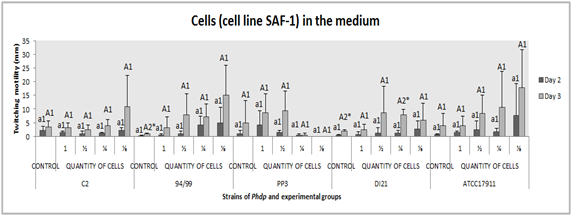
Figure 8 Effect of presence of cell debris in the medium on twitching motility of the Phdp strains tested: C2, 94/99, PP3, DI21 and ATCC 17911.
Small letters: Compare “groups” after 48 h of incubation
Capital letters: Compare “groups” after 72 h of incubation
Numbers: Compare “days of incubation” of each group
*Indicates that there are statistically significant differences
For various reasons, many authors have reached the conclusion that low and high temperatures alter fimbriae expression;29,38,39 bearing in mind, of course, that the meaning of low or high will depend on the bacteria (and its genetics). In our work, the best performance was obtained between 22 and 25˚C. At lower temperatures, especially at 18˚C, bacteria did not always grow and develop displacement. Studied the adhesion of a marine bacterium to surfaces and the number of attached bacteria was proportional to the density of suspended bacteria at 12˚C to 14˚,40 on the other hand, at 25˚C, only a few bacteria were attached to the surface, despite a high culture density. Nevertheless, we did not measure the number of bacteria, but the displacement on the solid surface. As stated in the introduction of this article, temperature and media affect fimbria expression; this was observed while studying type 1 fimbriae in Escherichia coli K-12 and these effects appear to be independent:41 low temperatures slow down even shut down the fimbriae synthesis because that affects the promoters switching and fim genes; for E. coli, the optimal temperature seems to be between 37 and 41˚C, however, these temperatures are too high for Photobacterium damselae subsp. piscicida and 28˚C, which is the lowest temperature tested, is still high.
Expression of fim genes is slightly lower in alkaline and acidic environments than at neutral pH, where it is maximal. Acidic environments also affect the transcription of fim promoters.33 So, this could explain the results obtained in this experiment. Twitching displacement happened at pH 7.0. At other pH values, it was only and occasionally observed in two strains, after three days of incubation and considerably fewer compared to that developed at neutral pH. But about the adhesion ability, the results achieved with an unidentified motile Gram-negative rod was no difference at pH 7.4 and 8.640 and with Vibrio spp. were a maximal adhesion at pH 8.2-8.5.42 The repressing effect on transcription of these genes at low pH is increased when osmolarity is moderate to high in Uropathogenic Escherichia coli;33 but Photobacterium damselae subsp. piscicida is a marine fish bacterium and can tolerate high NaCl concentrations, so it could be expected that its performance would be different.
Fimbriae expression depends on specific growth rate, which is independent of the growth-limiting nutrient used.29 On the other hand, in this study, the quantity of nutrient in the medium was reduced, observing perhaps a slight and not significant decrease in twitching displacement of strains 94/99 and DI21 at low concentration of nutrients. Nevertheless, with all the data, we think we can say that nutrient limitation does not seem to significantly affect phenotypically. In the study of Fletcher and Floodgate,41 the peptone concentration did not affect the number of attached bacteria either, even if the levels used were greater than the natural organic nutrient concentrations in seawater. On the other hand, Gally et al.41 observed that fimbria synthesis was slower in defined rich medium than minimal medium and can vary and be control by levels of aliphatic amino acids (alanine, isoleucine, leucine, valine).
No twitching displacement in NaCl-free culture media can be explained by the necessity of NaCl for Phdp to grow and accordingly, to move. This result contrasts with others, where an increased osmolarity reduces fimbriae expression;33 however, the adhesion of Vibrio spp., other marine bacteria, to mucus-coated glass slides is higher when the bacterial cells are incubated in seawater (3.5% NaCl) than in saline solution and PBS.42
Fimbriae production by bacteria is greater in broth than agar (1.5%) cultures38 and therefore, displacement is better on rather soft and wet surfaces (0.3% agar) than on relatively firm and dry surfaces (1.5% agar).43 We agree with this statement. Nevertheless, Liu et al.44 observed a better twitching displacement in 1.2-1.6% agar plates after testing agar concentrations ranging from 0.8 to 2.0%.
Slightly greater displacement was got on rough surface, it is known that such surfaces favour the biofilm formation;45 after all, bacterial adhesion is affected by physic-chemical properties of both the surface and the bacteria cell wall, including its structural features,46-48 in this case, fimbriae. Also, Anselme49 showed a relationship between roughness and adherent bacteria number on polyurethane-coated glass plates preconditioned in seawater. Whitehead et al.50 & Campoccia et al.51 studied this subject by testing different pits with specific diameter and depth, whereas in our assay, the marks were irregular on the plates and the results expressed as displacement and not bacteria number; in the first study, there was an increase of adherent bacteria number on rough surfaces, especially on the bottom of crevices,52 although the lowest number was obtained on the larger pit, but in the second one, no differences were observed. In addition, some authors have proposed that bacterial response to nanometer scale roughness is mediated by structures such as fimbriae.48,49
Adding metals to the medium (Ca, Co, Cu, Fe and so on),53 exposing bacteria to tissue cells it interacts with and feels tropism towards54 as our work, are some methods employed in order to study and promote cell adhesion and twitching displacement. That is it, Phdp strain PP3 showed a slightly better displacement in culture media with a lot of cells; strains 94/99, DI21 and ATCC 17911 too, but a lower amount of added cells was enough and better; however, for the strain C2, media with or without cell debris did not make difference.
In conclusion, according to the results, the “new” composition of the “twitching displacement” medium could be as follows: 3.7% (w/v) BHIB supplemented with about 1.0% (w/v) NaCl and 0.2% (w/v) agar, with a final pH of 7.0, and maybe, scrape petri dishes and add debris of SAF-1 cells 1:2-factor dilution to medium. If bacterial strains are incubated at 22-25˚C for three days, a good response should be achieved in terms of fimbriae production. Now, we must continue with the study of these pili-like structures recently discovered in Photobacterium damselae subsp. piscicida and their relationship to the ability of the bacteria to adhere and invade non-phagocytic cells, and their possible involvement in the development of immunity against Photobacterium damselae subsp. piscicida and the possible application of the same in vaccine formulations or as a therapeutic target.With this medium we can increase significantly the expression of such pili-like structures to work.
Dr. Acosta thanks to Fundación Ramón Areces for support this work by the Grant XVII Concurso Nacional para la adjudicación de Ayudas a la Investigación en Ciencias de la Vida y de la Materia (CIVP16A1810). Belinda Vega thanks to University of Las Palmas de Gran Canaria for suport with phD grant.
The author declares no conflict of interest.

©2017 Belinda, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.