Journal of
eISSN: 2469 - 2786


Research Article Volume 8 Issue 2
Centre for Interdisciplinary Research in Basic Science, Jamia Millia Islamia - A Central University, India
Correspondence: Fareeda Athar, Associate Professor, Centre for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia - A Central University, New Delhi-110025, India
Received: June 22, 2020 | Published: July 6, 2020
Citation: Beg MA, Thakur SC, Athar F. Computational annotations of mycobacterial Rv3632 that confers its efficient function in cell wall biogenesis. J Bacteriol Mycol Open Access. 2020;8(2):46-53. DOI: 10.15406/jbmoa.2020.08.00272
Mycobacterium tuberculosis (Mtb), an etiological agent is a key pathogen responsible for devastating disease Tuberculosis (TB). Development of drug resistant strains and failure of BCG vaccine prompts us to develop a new therapeutic regimen. Moreover, Mtb practices a variety of machineries for avoiding its eradication by an infected host; and therefore, practical substitutions for inhibiting the host-pathogen interaction are thus required regularly. As an essential cell wall component, this manuscript updates our basic existing knowledge of Rv3632 gene, a transmembrane protein and a probable contributor of biosynthesis of the mycobacterial cell wall. The methodology used for updating the information was purely bioinformatic analysis which includes its sequence annotation, structure modelling and validation, molecular docking with already existing cell wall inhibitor and finally mutational analysis. Results showed that Rv3632 comprise of several transmembrane helices that range with different amino acid chains. Ab initio modelling was used for structure prediction which was validated by SAVES metaserver and data confirms 88.5% residues presence in the allowed region. Cell wall biosynthesis inhibitors were used to target Rv3632 due to its prediction in involved in cell wall synthesis. The docking score was as Ethionamide (-4.4), Thioacetazone (-4.4), Prothionamide (-4.3), Cycloserine (-3.7), Secnidazole (-3.4), Ethambutol (-3.4) and Metronidazole (-3.2). The study was ended by mutation analysis at TRP (W31) and TYR (Y71) and the result shows that substitution by glycine at both places decreases protein stability the most. This study enlightens the fact that mycobacterial cell wall targeting drug tolerance is majorly due to internal point mutations at specific residues. Thus, this report enhancing the understanding of molecular mechanism and components involved in mycobacterial cell wall synthesis and help in targeting those components and opens the way to novel therapeutic development.
Keywords: Rv3632, Ab initio modelling, molecular docking, point mutation, structural stability
TB, tuberculosis; Mtb, mycobacterium tuberculosis; MDR-TB, multidrug-resistant TB; PDB, protein data bank; NCBI, National Center for Biotechnology Information; kD, Kilodalton; TMRs, transmembrane helices; 3D, three dimensional; TRP, tryptophan;TYR, tyrosine
Mycobacterium tuberculosis is the causative agent of tuberculosis which is one of the highest and frequent infectious diseases. This disease has its ancient origin and now coevolved with the evolution of the Homo sapiens species.1 In human primates this bacterium can retain in the surrounding environment of several host immune cells.2 Mycobacterium maintains highly repetitive life span which possess a range of heterogeneous origin and physiological state many of which has a difference from other bacterial species.3 The symptoms of this disease can be ranged from a latently non-transmissible disease to severe transmissible disease. In case of latently infected individuals, this bacterium gets success in escaping the host immune system and resides in human alveolar macrophages for a long period time.4,5 But in case of actively propagating disease, these latently reside bacilli bring out from the macrophages in which they form a granulomatous lesion and move to the lymphatic vessels as well as other parts of the body.6 Granuloma is the modified host immune environment which develops in response to the bacterial infection. This structure establishes a mutual relationship between host and bacterium because both parties obtain equal benefits from each other.7‒10 Rv3632 peptides being distinguished as promising parts of a multi-epitope antibody coordinated against mycobacterial cell interacted with the host. The requirement for covering the best measure of mycobacterial access courses to infection of target cells and for delivering a viable immunogenic response considered. Rv3632 is present in Mtb H37Rv cell wall. Various studies distinguished by mass spectrometry in the proteomic investigation in concentrates got with Triton X-114 and has been perceived by mass spectrometry in protein layer portions and complete mycobacterial cell lysate, however not in culture filtrate. Transposon site hybridization (TraSH) investigation has announced that the quality encoding this protein is neither essential for mycobacteria nor important for mycobacterial in vitro development.11 The putative role of Rv3632 is its participation in arabinogalactan biosynthesis which is a major component of the mycobacterial cell wall and might be involved in its pathogenesis. Its direct role is predicted to be in the stabilization of polyprenyl-phospho-N-acetyl galactosamine synthase (PpgS) that leads to polysaccharide biosynthesis.12 As this protein is transmembrane therefore it might be involved in mycobacterial virulence and its attachment to host surface. This manuscript involved essential aspects of Rv3632 promising facts about generating multi-epitope antibody against mycobacterial cell involved with Host-pathogen interaction which is foremost the major part of having an infection and thus studying how to inhibit this interaction is an important aspect in controlling infection of this pathogen. Of note, this manuscript highlights various aspects of Rv3632 which is also demonstrated by the flowchart of computational approaches in Figure S1, these tools/servers help for the predicting role of this gene in adhesion of the bacterium to the host cell and thus helpful in providing essential information useful for preventing infection and decreases mortality due to this disease.
Complete genome sequence of M. tuberculosis H37Rv is available in the National Centre for Biotechnology Information (NCBI).13 In this manuscript, we used several tools for specific functional studies in respect of Rv3632 from M. tuberculosis H37Rv details in (Figure S1). Here now, the computational methods framework consists of retrieval of the sequence, prediction of transmembrane region, Signal peptide analysis, disordered region, ab initio modelling, molecular docking and mutational analysis.14,15
Transmembraneregions orientation
Studies of transmembrane helices (TMRs) are significant in the context of analyzing the membrane protein. To evaluate the TMHs prediction there are many algorithm-based tools already present i.e. Phobius, SOSUI, TMHMM and HMMTOP. Phobius sever predicted the transmembrane topology and signal peptides for using the FASTA format amino acid sequence of a query protein.16 SOSUI server had been used for discriminating between the membrane proteins and soluble proteins based on an amino acid sequence of the query protein. The accuracy of the classified protein was 99% and the corresponding value for the TMHs prediction was 97%.17 TMHMM server is based on a hidden Markov model algorithm which predicts the probable location and orientation of the transmembrane helices. TMHMM server output the sequenced labelled as inside or outside and predicted that it contains transmembrane helix.18 HMMTOP server is an automatic server which predicts the localization of the TMHs and topology of the protein by using the amino acid sequence of a protein.19 For interactive protein data study was done by Protter, it’s a web-based tool which supports to given visualizing hypothesis generation by using the amino acid sequence in the context of protein topology. Protter supports UniProt protein accession and numerous protein sequence, which can be readily extended via modular plugins. Protter visualizations sequence datasets show the integrated visual analysis of membrane proteins and peptide selection for targeted proteomics.20
Protein sorting prediction
The protein sorting prediction of cleavage site was done by SignalP 3.0 server. Rv3632 protein sequence was examined by SignalP 3.0 server. The Y and C score distinguishes between in peaks by choosing the one where the slope of the S-score is the step. The Y-score (combined cleavage site score) and S-score (signal peptide score) differentiate in positions within the signal peptides from positions in the established part of the proteins and proteins deprived of the signal peptides and the C-score is trained to be high at the position proximately after the cleavage site.21,22
Prediction of disordered region
One more step to validate the protein topology analysis prediction of the disordered region by using different online servers like DisEMBLTM, IUPred2A and PONDR etc. By using PONDR online tool we predicted the intrinsic order/disorder from the amino acid sequence by VSL2, VL3 and CaN-XT method. The sequence-structure relationships indicate that disorder is an encoded property, and the predictions strongly suggest that proteins in nature are much richer in intrinsic disorder than are those in the Protein Data Bank.23,24
Ab initio protein modelling
Three-dimensional (3D) protein modelling of the protein from its amino acid sequence remains unsolved if the query protein has PDB homolog of known structure then it will be easy to model the query amino acid sequence.25 Rv3632 sequence was retrieved from the Mycobrowser database and three-dimensional (3D) modelling was predicted by the ab initio modelling server QUARK online server which constructs the 3D model of a protein from an amino acid sequence. This online server built a model from a small fragment (1-200) residue long by the Monte Carlo simulation under the guide of atomic-level knowledge-based force field. This server does not have a homologous template in the PDB library.26,27 The Rv3632 protein model was validated by using SAVES (Structure Analysis and Verification Server) metaserver and protein quality estimation by using ProQ server which has LGscore and MaxSub which define the model is fairly good, very good and extremely good.28‒33 The 3D structure was modelled (sequence length 1-114) and refined by the 3Drefine server. This server follows the two-step first optimization of hydrogen bonds and second energy minimization by MESHI molecular modelling package.34
Molecular docking
The topological properties of protein structures, including binding pockets was determined by using CASTp 3.0. Few residues are responsible for predicted ligand binding site in the protein where the ligand can reversibly bind.35,36 Interaction analysis of the protein and ligands done by AutoDock Vina38 and PyRx 0.8 (FastSpring, Amsterdam, Netherlands) were produce the dock file PDBQT and docking analysis where predicted binding score or binding affinity (kcal/mol) and both ligand-receptor dock file visualized by using PyMOL and for interaction analysis was done by using Discovery Studio visualizer.37‒40 Rv3632 was modelled by ab initio modelling which contains a peptide81 TLSTYMRFKDLELRYAIAR100 which contains the specific binding to infection target cells A549 and U937.11 Preparation of the receptor molecule was initiated with the removal of water molecules and add polar hydrogen only then convert into PDBQT by using AutoDock Vina which is followed by grid formation.41 The ligands that target cell wall synthesis are drugs Ethionamide, Prothionamide, Thioacetazone, Metronidazole, Secnidazole, Ethambutol and Cycloserine downloaded by PubChem online server by using PyRx 0.8. All the ligands were in SDF file, then we minimize the energy which is favourable for converting into PDBQT file.42,43 The interaction analysis of the protein-ligand and interaction of the binding sites and the possible dock conformations of the 2D ligand-receptor interactions by using Discovery Studio.40 Further, binding analysis by using visualization approach was carried out to understand the binding pattern of the drug with proteins.
Mutational analysis
The mutational consequence on protein by single point mutation is an important phenomenon but how to select the residues for point mutations is a big question. However, Insilico analysis provides this information in EASE-MM server.44 Ease-MM is based on support vector machine (SVM) models generated a Pearson correlation coefficient of 0.53–0.59 in the validation of 10-fold increase and free testing. At the point when distinguished with structure-based energy capacities, EASE-MM accomplished an equivalent or better execution has given values predicted the changing stability of single point mutation via protein sequence. EASE-MM stands for Evolutionary Amino acid and Structural Encoding with Multiple Models which is based on support vector machine (SVM) models generated a Pearson correlation coefficient of 0.53–0.59 in the validation of 10-fold increase and free testing. At the point when distinguished with structure-based energy capacities, EASE-MM accomplished an equivalent or better execution.45,46 Structure stability changes upon point mutation by using DynaMut webserver benefitted in two manners; one can be utilized to decide and to contemplate protein elements. Subsequently, it may be utilized to survey the effect of single point change. The cut off worth set for this server is ΔΔG≥0 as stabilizing and ΔΔG< 0 as destabilizing. Normal Mode Analysis (NMA) is a computational strategy that approximates the dynamic practicality of a structure around an adjustment through consonant development. This has been used to make possible improvements and appropriately give beneficial bits of information into protein developments, and their accessible conformational assortments.47,48
Transmembrane helix predicted by using the Phobius, SOSUI, TMHMM and HMMTOP servers. Phobius server result showed the topology of the domain (TOPO_DOM) and transmembrane helices (TRANSMEM). TOPO_DOM region of the 1st-5th and 49th-67th was non-cytoplasmic else 24th-29th and 89th-114th were cytoplasmic.16 TRANSMEM region is in 6th-23rd, 30th-48th and 68th-88th. SOSUI server analysis was done to evaluate protein hydrophobicity if it exists this server makes a label that residue of this protein as a transmembrane region. SOSUI server describes the transmembrane region as 1st-21th, 29th-51st and 65th-87th.17 TMHMM sever result show that in Rv3632 protein, there were 3 transmembrane helices (TMHs) 4th-21th, 30th-48th and 68th-85th. The predicted sequence as outside region 1st-3rd and 49th-67th, the inside labelled sequence is 22nd-29th and 49th-67th.18 HMMTOP server result had been showing that there were 3 transmembrane helices on 4th-20th, 31st-48th and 69th-86th amino acid residues the output of the total entropy of the model is 17.0013 and the best path of the entropy is 17.0017.19 For visualizing hypothesis of transmembrane helices (TMRs) by using amino acid for protein topology shows Rv3632 is a membrane protein which consists three TMRs on position 6th-21st, 30th-48th and 68th-87th which is shown in Figure 1.20
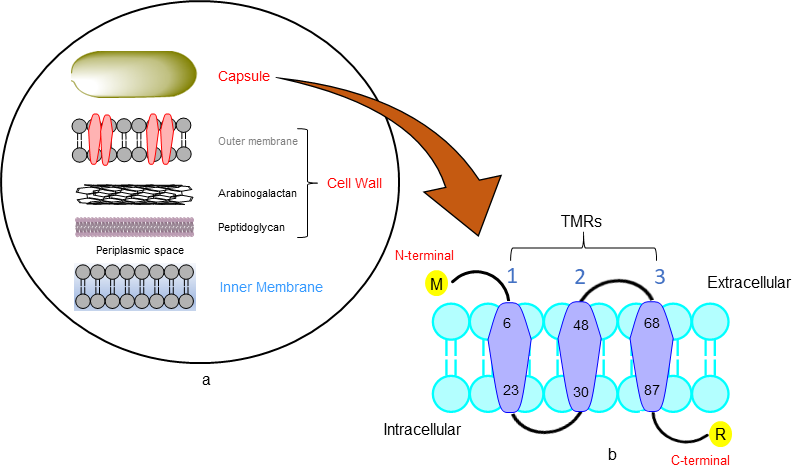
Figure 1 Mycobacterial membrane (A) The different component of the mycobacterial cell envelope based on the description Chen et al.12 (B) Protter server showing the visualizing hypothesis of trans membrane helices (TMRs).
Protein sorting prediction
Prediction of the signal peptide cleavage site, the phenomenon is also known as protein sorting was done by SignalP 3.0 online server where study the organism group is mainly for Gram-positive bacteria with the D-score cut-off value of 0.45. In Rv3632 incisive prediction of signal peptide result outcome shows that there was 27th number amino acid residue with highest Y-Score and therefore the other two cleavage sites with relatively lower Y-Score predicted to be as 25th position residue. Most likely cleavage sites were predicted between 26 and 27th position towards the N-terminal of serine as shown in Figure 2.22
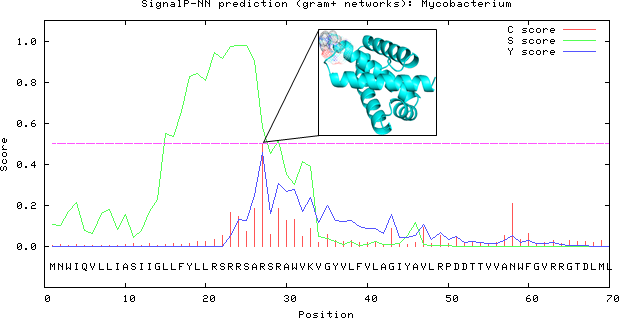
Figure 2 Predicted cleavage sites done by SignalP 3.0 server which showing the location of most probable cleavage sites 26th and 27th residue at highest Y-Score on 25th position represents as cartoon model.
Prediction of disordered residues
In Rv3632, the disordered residues prediction was done by PONDR by using VSL2, VL3 and CaN-XT plots shows their greater than 0.5 scores were disordered residues and scores less than 0.5 were predictions of ordered residues. The PONDR plot shows in Rv3632 the VL3 and CaN-XT plots were residues of the ordered region except for VSL2. Here residue number 24-27 and 107-114 scores above 0.5 which means these residues predicted as in the disordered region. Comparison of disorder residues prediction between PONDR-VSL2, PONDR VL3 and PONDR CaN-XT plot is shown in Figure 3.23,24
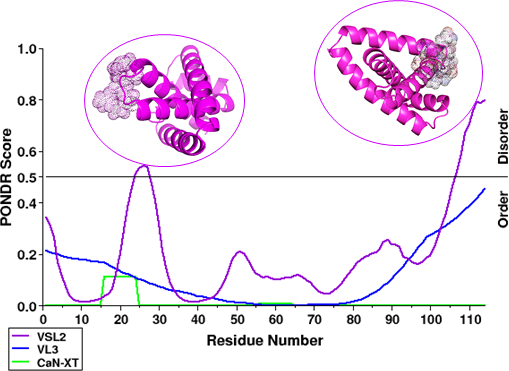
Figure 3 Comparison in graph plot for disordered protein in between VSL2, VL3 and CaN-XT plots, where scores greater than 0.5 are predictions of disordered residues. In graph plot represent regions (from N- to C- termini) in VSL2 plot residue no. 24-27 and 107-114 in protein cartoon model dot represents disordered region.
Ab initio protein modelling
Three-dimensional structure of Rv3632 from M. tuberculosis was modelled by QUARK server using ab initio method which constructs a model of correct fold 1/3 cases of short protein which have ≤200 amino acid residues. Using QUARK server there were five structures where we found the best predicted model. After all the five predicted protein model validation was completed by SAVES metaserver, its sub server like Verify 3D, ERRAT and RAMPAGE predicted model 2 has a best score as shown in Figure 4.27,28 In model validation Verify3D has 78.95% score, ERRAT has 87.7358 score and RAMPAGE where analysis the Ramachandran plot which showed the allowed and disallowed residues showing the predicted model of Rv3632 has 88.5% residues are in the allowed region. In predicted model quality prediction has LGscore is 6.092 and MaxSub is 0.328 which defines this modelled has satisfactory validation.29-32
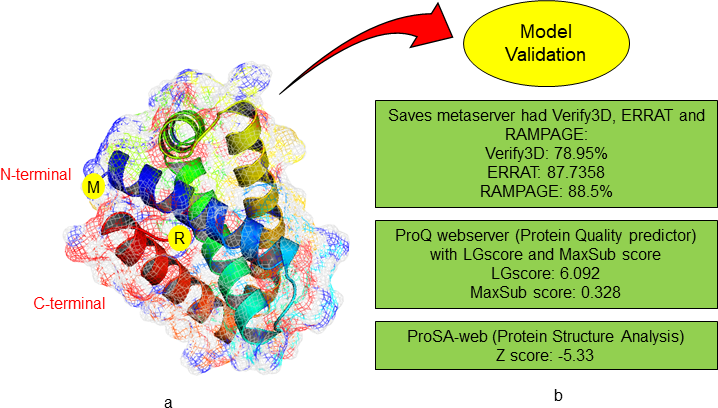
Figure 4 Protein modelling using QUARK server (A) Rv3632 model represents in mesh surface cartoon model. (B) Predicted model validates by using SAVES, ProQ and ProSA-web servers.
Molecular docking of Rv3632 with current TB drugs: target cell wall synthesis
Molecular docking studies give the precise orientations in between receptor and compound. Due to drug resistance, TB is more infectious, multidrug resistant (MDR)-TB and extensively drug resistant (XDR)-TB the drugs given for the treatment of this disease are no longer capable of curing the infection due to the resistance nature of this bacterium toward drugs. For receptor preparation, the grid parameter was saved in CONF file which is centre X-58.831, Y- 58.777 and Z-58.208 and size X-16, Y-12 and Z-14 used for docking.36‒38 By using molecular docking, we used current TB drugs involve in cell wall synthesis which was having docking scores Ethionamide (-4.4), Prothionamide (-4.3), Thioacetazone (-4.4), Metronidazole (-3.2), Secnidazole (-3.4), Ethambutol (-3.4) and Cycloserine (-3.7) against Rv3632 showing in Figure 5. The interaction analysis of Rv3632 complexes suggested that active sites contain many significant residues to which hydrogen bond interaction on Trp31, Phe77, Thr127 and Thr84 present in active sites and substrate binding domains as shown in Figure 6. This interaction studies show complex Rv3632_Ethionamide and Rv3632_Thioacetazone has highest docking score and residue Trp31 has common in all drug binding pose37-41 and therefore further mutation analysis at this residue was done to confirm whether this mutation destabilizes protein or not?
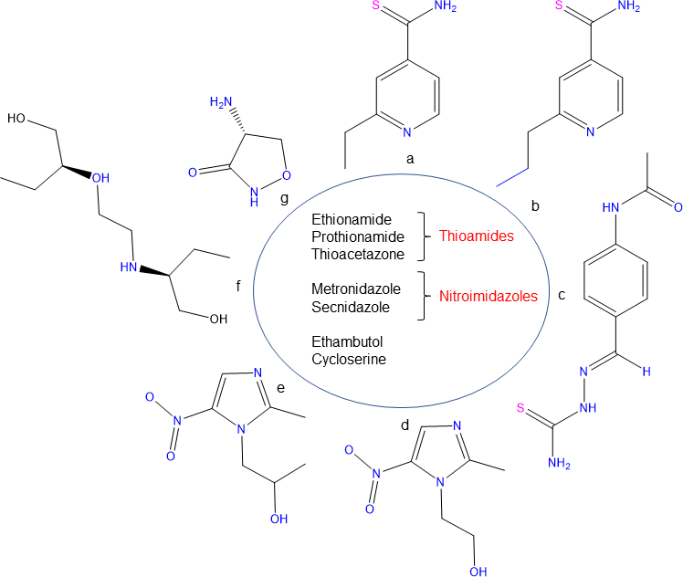
Figure 5 Structures of TB drugs involved in their cell wall synthesis (A-G) Ethionamide, Prothionamide, Thioacetazone, Metronidazole, Secnidazole, Ethambutol and Cycloserine.

Figure 6 2D interaction analysis of TB drugs against R3632 the complex interaction between ligand receptor (A-G) Ethionamide, Prothionamide, Thioacetazone, Metronidazole, Secnidazole, Ethambutol and Cycloserine.
Point mutation changes structural stability
EASE-MM server predicted the highest stability change in residue by the sequence based method and DynaMut server predicted the mutated amino acid changes in protein structure regarding protein stability.44 EASE-MM server comprises of five specialized models of support vector machines and then select final prediction based on selected secondary structure model and its surface area. After EASE-MM server predicted conclusion selected residues were Trp (W31) and Tyr (Y71). EASE-MM predicted all the consequence of mutation with W31 and Y71 in Figure 7. Glycine (G) has highest negative score rather than other residues which means more stability decreases of the protein for W31 (-3.0435) and Y71 (-4.447) was occurred by mutating these residues to Glycine as shown in Figure 7.45,46 Impact of the point mutation was also studied by using DynaMut server which had two diverse ways, (1) study of protein dynamics and (2) effect of point mutation on protein structural stability. In this server, mutation study includes three different servers mCSM, SDM and DUET which results for W31 has mCSM (-3.656), SDM (-2.620) and DUET (-3.373) and Y71 has mCSM (-3.072), SDM (-2.320) and DUET (-2.964).38 DynaMut server predicted both residues W31 and Y71 point mutation has a negative value which shows the destabilizing effect.39,40 Effect of interatomic interaction on the single amino acid mutation predicted the drastic loss in Interatomic interactions. All hydrogen bonds, halogen bonds and hydrophobic interaction were disrupted as shown in Figure 7.47,48
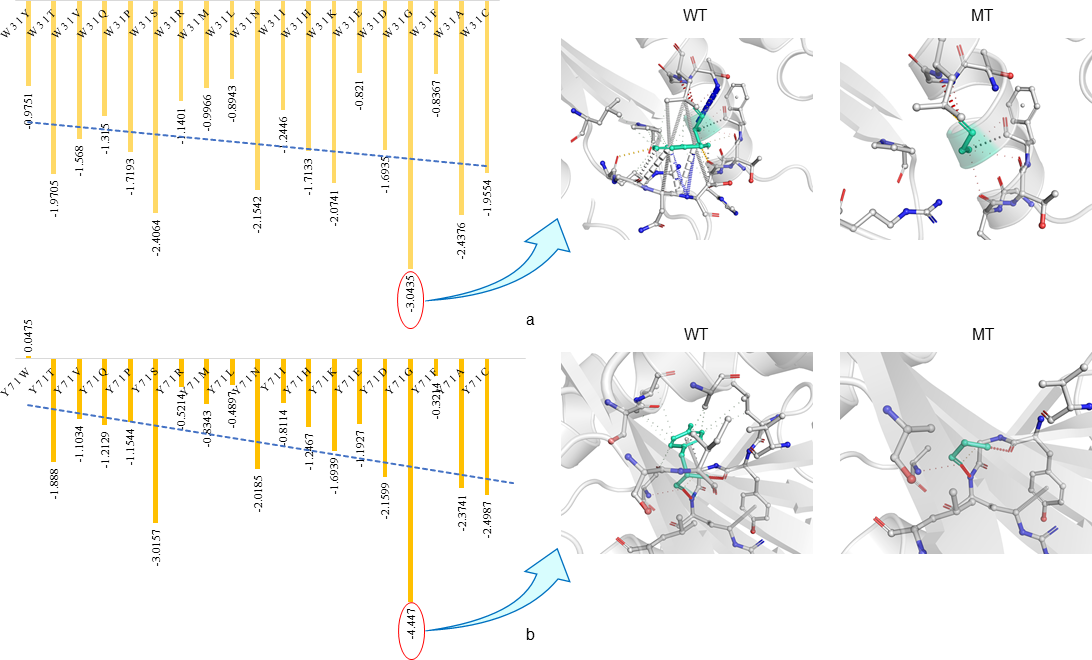
Figure 7 In Rv3632 predicted point mutation stability changes by using EASE-MM and DynaMut (A) EASE-MM server predicted the W31 and Y71 stability changes resulting Mutation with Glycine decreases stability in both. (B ) DynaMut visualize the drastic changes in interatomic interaction between wildtype (WT) and mutant (MT) which shows mutation showing the bond disruptions.
In concluding the overall view of the manuscript, we can notice that Rv3632 is an important integral part of the mycobacterial cell membrane. It is crucial for the efficient functioning of PPGS and thus play an important role in cell wall biosynthesis. Mycobacterium tuberculosis is an efficient bacterium in terms of its infection intensity. It employs different genomic mutation to acquire consistent drug resistant feature and therefore we are facing an urgent requirement of therapeutic drug or vaccine against this disease. This manuscript pointed on essential aspects of Rv3632 that which is essential in establishing the experimental design of drug designing methodology. Of note, Rv3632 is a transmembrane protein. Ethionamide, Prothionamide, Thioacetazone, Metronidazole, Secnidazole, Ethambutol were used for docking analysis. These drugs were prioritized for their action against the mycobacterial cell wall and thus these were used here as a control agent to check their binding affinities with the protein. The important findings can be concluded as the order of docking score as Ethionamide > Thioacetazone > Prothionamide > Cycloserine > Secnidazole > Ethambutol > Metronidazole and interaction analysis shows TRP31 amino acid is present in all complex structures. Further mutational analysis at TRP (W31) and TYR (Y71) found that mutation with glycine shows a large decrease stability. This report emphasizes a novel site about Rv3632 which is important for providing a path to understand its proper functioning that enhances targeting this protein by a therapeutic drug or developing a drug against this protein.
Md Amjad Beg: Conceptualization idea, Methodology designing, Software handling, writing manuscript and evaluate results; Sonu Chand Thakur: Reviewing; Dr Fareeda Athar: Reviewing and corresponding author.
The authors acknowledge support from the Centre for Interdisciplinary Research in Basic Science, Jamia Millia Islamia University. Md Amjad Beg also acknowledges UGC-MANF (MANF-2017-18-UTT-88071) for the financial support and Jamia Millia Islamia University.
The authors declare that they have no potential conflict of interests.

©2020 Beg, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.