Journal of
eISSN: 2469 - 2786


Research Article Volume 9 Issue 1
1Institute of Soil and Water Resources and Environmental Science, Zhejiang University, China
2Department of Microbiology, Kohat University of Science and Technology, Pakistan
3The China-US (Henan) Hormel Cancer Institute, China
4Department of Biotechnology, Bacha Khan University, Pakistan
5Institute of Biotechnology, Zhejiang University, China
6Department of Chemistry, Kohat University of Science and Technology, Pakistan
Correspondence: Muhammad Afzal, Institute of Soil and Water Resources and Environmental Science, College of Environment and Resource Sciences, Zhejiang University, Hangzhou 310058, China
Received: February 11, 2021 | Published: March 17, 2021
Citation: Afzal M, Saeed M, Muhammad N, et al. Characterization, antibiotic sensitivity assay and phylogenetic analysis of bacterial isolates of river water. J Bacteriol Mycol Open Access. 2021;9(1):7-13. DOI: 10.15406/jbmoa.2021.09.00289
Objective: Rivers and streams are less studied area with respect to its microbial ecology. This study was conducted to isolate, physiochemical characterized and analyzed phylogenetically bacteria of River Swat, Pakistan and to investigate the antibiotic sensitivity of these isolates.
Methods: Total 65 water samples were collected from five different sites, named Charbagh, FizaGatt, Kaladeer, Barikot and Khaiali in Charsadda, Pakistan. Water samples were taken from shore, middle and from the mud of the river in each site. The bacteria were identified by biochemical tests and confirmed at species level by PCR. Resistance to various antibiotics was determined by disk diffusion method.
Results: Total of 23 bacterial isolates was isolated, of which 18 isolates were found as Gram-negative rods. Most of the isolates grown at pH range 5-9 and grow best at salt concentration ranges from 0-5%. Maximum growth was observed at 35°C. The isolates exhibited different levels of resistance to various antibiotics, with the highest resistance (95.54%) to Ampicillin. In addition the least percentage of the isolates (31.81%) showed resistance to Ciprofloxacin and for rest of the antibiotics, no significant difference in resistance reaction was observed. The highest number of bacterial isolates (31.81%) showed an intermediate reaction against Ciprofloxacin The 16S rRNA gene sequencing analysis showed the phylogenetic relationship among bacterial isolates of river Swat at different sites.
Conclusion: The presence of antibiotic-resistant isolates in river water may cause water-borne diseases. The isolates have a great biotechnological importance and need further exploration.
Keywords: river swat, waterborne bacterial diseases, antibiotic resistance, phylogenetic analysis, microbiology
Water is essential for life and is required for proper body function. Availability of good quality drinking water is essential human life.1 More than 97% of the global water is salt water, and only 3% is freshwater,2 out of which 69% of the fresh water is used for domestic purpose, 22% is being used in industries, 8% is used for irrigation.3 About 2% of the total volume of fresh water is contained in the lakes, while rivers account for a very small amount (0.0001%) of water on the earth surface.4 In rural areas, the main sources of drinking water are dams, wells, rivers, streams, ponds that have been reported to harbor waterborne pathogens.5 The river is a natural watercourse, whose characteristic feature is a continuous movement of running water. Rivers and streams are less studied area with respect to its microbial ecology.6 In river ecosystems, there are free-swimming bacteria as well as bacteria attached to the mobile interfaces of aquatic aggregates along with algae, fungi, and protozoa that constitute a major part of the mixed aquatic ecosystems.7 In fresh water, besides Bacillus, Monococcus,and Micrococcus, other bacterial genera, including Pseudomonas, Caulobacter, Achromobacter, Flavobacterium, Alcaligenes, Thiobacillus, Leptothrix and Clostridium are reported to be present.8 District Swat (35°23′N 72°11′E) lies in Khyber Pakhtunkhwa Province of Pakistan. River Swat starts from the Hindu Kush range, flows through the Kalam valley and plays an important role in the economic life of the valley. The water flow is useful for human existence, industrial, agricultural and domestic purposes. The last decade has seen significant influence of society on environment due to unique intensification in population growth and hasty rate of urbanization as well as the escalation of the use of fragile and marginal ecosystem. Several studies reported that urbanization increase the concentration of various nutrients such as ammonia, phosphorous, nitrogen and nitrates that results pH variations. The urbanization also results discharge of pathogenic bacteria into water environment. The determination of these bacterial isolates has been great contest to microbial ecologists. This study was therefore aimed to characterize bacteria, isolated from river Swat, Pakistan and to investigate the antibiotic sensitivity of the bacterial isolates.
Water samples were collected in a sterilized falcon tube from five different sites at 1m depth and were kept at 4°C in a refrigerator.
Isolation and characterization of bacterial isolates
Water samples were poured aseptically on Tryptic soy Broth and Ager (TSA) (Difco) and the plates were incubated at 28°C. After 24 hours, different colonies were observed. The bacterial isolates were characterized by Gram’s staining, followed by cell and colony morphology. Bacterial isolates were tested for their growth at different pH levels (4-11), temperature ranges (0, 10, 28, 35 and 40°C) and NaCl concentrations (1-20%) on TSB medium. All bacterial isolates were optimized to pH in the range of pH from acidic to basic.
Antibiotics sensitivity test
The Kireby-bauer method was used for antimicrobial susceptibility test.9 The inoculum was prepared by making a direct broth or saline suspension of isolated colonies from 18-24 hours nutrient agar culture plates. The suspension was adjusted to match 0.5 McFarland turbidity standard, using saline and vortex mixer. Optimally, within 15 minutes after adjusting turbidity of the inoculums suspension, a sterile cotton swab was dipped into the adjusted suspension. The swab was rotated several times and pressed firmly on the inside of the wall of the tubes above the fluid level to remove excess inoculums from the swab. The dried surface of Mueller-Hinton agar plates was inoculated by streaking the swab over the entire surface. The procedure was repeated by streaking two more times, rotating the plate approximately 60° each time to ensure equal distribution of inoculums. As a final step, the rim of agar was swabbed. The lid was left open for 3-5 minute, to allow for any excess surface moisture to be absorbed before applying the drug impregnated disks. The antimicrobial discs were dispensed onto the surface of the inoculated agar plate. Each disc was press down to ensure complete contact with agar surface. The plates were kept inverted in an incubator at 30°C for 24 hours (Supplementary).
Analytical profile index 20E
This test was used for identification of the isolates at the species level. An incubation box was prepared by adding about 5ml sterilized water to make a humid environment. To standardize the Inoculum density for a susceptibility test, a BaSO4 turbidity standard, equivalent to a 0.5 McFarland standard, was used. A BaSO4 0.5 McFarland standards were prepared by adding a 0.5ml aliquot of 0.048mol/L BaCl2 (1.175% w/v BaCl2.2H2O) to 99.5ml of 0.18mol/L H2SO4 (1% v/v). The absorbance at 625nm was adjusted at 0.008 to 0.10 for the 0.5McFarland standards. The BaSO4 suspension was transferred in 6ml aliquots into screw-cap tubes of the same size as those used in growing or diluting the bacterial inoculum. These tubes were tightly sealed and stored in dark at room temperature. The bacterial suspension was prepared in 5 ml normal saline and the isolated colonies were taken. The bacterial suspension was compared to 0.05% McFarland standard. By using a sterile syringe all wells of the strip were filled according to the instructions of API 20E manual. An anaerobic environment was created in tubules, by putting a drop of mineral oil overlay and the box was incubated at 37°C for 24 hours.
PCR amplification and sequencing of 16S rRNA gene
To amplify the 16S rRNA gene the genomic DNA was extracted according to.10 Briefly, few isolated bacterial colonies were suspended in TE buffer in a microcentrifuge tube, followed by heating for 10 min at 95°C and centrifugation at 6,000 rpm for 5min. The supernatant was used as template DNA for the amplification of 16S rRNA gene. The 16S rRNA gene of all the isolated isolates was amplified by polymerase chain reaction using primers 9F (5'-GAGTTTGATCCTGGCTCAG-3') and 1510R (5'-GGCTACCTTGTTACGA-3') using Premix ExTaq (Takara, Japan) following the protocol described previously.11 The PCR was carried out in ABI Veriti PCR Machine (Applied Biosystems, USA) using optimized PCR Program: initial denaturation at 94°C for 2 min; 30 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1 min and extension at 72°C for 1:30 min. The final extension was performed at 72°C for 5min. The amplified PCR products of 16S rRNA gene of bacterial isolates were purified and sequenced using the primers 27F (5'-AGAGTTTGATCMTGGCTCAG-3') and 1492R (5'-ACCTTGTTACGACTT-3') using commercial service of Macrogen Inc. Korea (http://dna.macrogen.com/eng/).
Phylogenetic analysis of bacterial isolates
Bio-Edit software12 was used to assemble the fragment sequences of 16S rRNA gene. Using 16S rRNA gene sequences, the isolates were identified by BLAST search on EzBiocloud Server). Closely related type species sequences were retrieved to assess the molecular evolutionary relations and to construct a phylogenetic tree using MEGA version 6.13 A phylogenetic tree was constructed from unambiguously aligned nucleotides using the neighbor-joining (NJ) algorithm. The relationship stability was evaluated by bootstrap analysis performed using 1000 re-samplings of the neighbor-joining data for the tree topology.
The optical density, pH, temperature and electric conductivity of the river water collected from the sampled areas (Table 1). Physical analysis of the water samples depicts that as the river passes through the populated area, an increase in optical density, electrical conductivity and temperature was observed. At Charbagh, neutral pH (7.04) was noted, but with the down ward flow of the river, its pH moves to alkalinity. At Khiali, Charsadda, where river Swat joins River Kabul, the pH of was noted as 8.08.
|
Sampled areas |
Sample codes |
OD |
pH |
Temp (°C) |
EC (µs/cm) |
|
|
Water |
Air |
|||||
|
Charbagh |
MMC |
1.034 |
7.04 |
13 |
20 |
0.073 |
|
Fiaza Gut |
MMF |
1.017 |
7.19 |
14 |
21 |
0.085 |
|
Kaladeer |
MMK |
1.065 |
7.43 |
15 |
25 |
0.085 |
|
Barikot |
MMBK |
1.350 |
7.70 |
21 |
28 |
0.112 |
|
Khiali, Charsadda |
MMKC |
1.678 |
8.08 |
22 |
42 |
0.137 |
Table 1 Physical analysis of river water
Gram’s reaction and growth conditions
Out of 23 bacterial isolates, 18 were found to be Gram-negative rods, while five isolates were observed as Gram’s positive cocci (Table 2). No growth was seen at 0°C, 4°C, 20°C. Maximum growth was observed at 35°C and moderate growth was observed at 28°C and 40°C.The NaCl tolerance was optimized for all the 23 isolates and fined their optimum growth on different NaCl percentage. The observed results of these isolates are shown (Figure 1). Nine of the bacterial isolates grow best at salt concentration 0%, one of the bacterial isolates (MMBK-DL) grows best at NaCl concentration 1%. Six of the bacterial isolates grow best at NaCl concentration 2%, 5 of the isolates grows best at NaCl concentration 3% and two of the isolates grow best at concentration 10%. Growth ranges of the 17 isolates were 0-13%, two of the isolates growth range was 0-11% and five of the isolates have their growth range 0-17% salt concentrations. All the 23 bacterial isolates were optimized to pH in the range of pH from acidic to basic. The optimum pH and their growth range of all the isolates are shown in the (Figure 2). All the isolates showed maximum growth at neutral pH 7-9. None of the isolates could grow at pH 4 as well as at pH 10.
Gram reaction |
Isolates |
Gram’s negative |
MMBK.Mb, MMBK-M2a, MMF3a, MMF1a, MMC3e, MMBK-Ma, MMKC, MMBK-Sm, MMC2a, MMF1b2, MMBK-Dl, MMBK-Dy, MMBK-Sc, MMBK-Sf, MMF1c, MMBK-Sd, MMF1a1, MMBK-SI |
Gram’s positive |
MMF1b3, MMBK-Dk, MCMs, MMF1b4, MMF3a2 |
Table 2 Gram’s reaction
Antibiotics sensitivity assay
Antibiotic sensitivity of the isolates against various antibiotics was checked by disc diffusion method and the results are represented in (Figure 3). The data depicted that maximum bacterial isolates (95.45%) were found resistant to Ampicillin, while the least percentage of the isolates (31.81%) showed resistance to Ciprofloxacin and for rest of the antibiotics, no significant difference in resistance reaction was observed. The highest number of bacterial isolates (31.81%) showed an intermediate reaction against Ciprofloxacin, while no isolate (0%) showed an intermediate reaction against Ampicillin. For the sensitive reaction, the maximum (45.45%) isolates were recorded as sensitive against Doxycycline, followed by Gentamycin, where 40.90% of the bacterial isolates showed a sensitive reaction. The least number of bacterial isolates (4.45%) showed a sensitive reaction against Ampicillin and Fusidic acid.
Analytical profile indexes
All the isolates were then tested on Analytical Profile Indexes (API) of BioMerieux France. Through API20E Pseudomonas spp, Proteus spp, Stenotrophomonas spp, Vibrio spp, Aeromonas spp, Salmonella spp, Serratia spp, Providencia spp, and Serratia spp were identified.
Phylogenetic analysis
The product after the amplification of 16S rRNA of bacterial isolates and subsequent Agarose Gel Electrophoresis are as shown in (Figure 4). Neighbor-Joining tree of the isolate MMBK-Sc, MMBK-M2a, and MMF1b2 with its closely related species, was constructed as shown in (Figure 5). Most of the 16S rRNA sequencing showed a maximum phylogenetic relationship with the Bacillus genera as shown in (Figure 6).
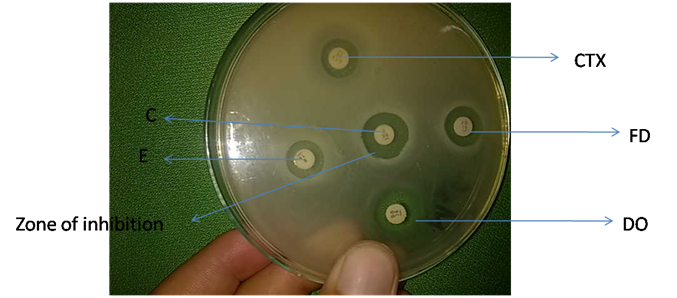
Figure 4 Antibiotic sensitivity test for MCMs, where C represents Chloramphenicol, CTX shows Cefotaxime, DO denotes Doxycycline, E represents Erythromycin and FD shows Fucidic acid.
Physiological characteristics
Bacterial isolates were tested for their growth at different pH levels (4-11), temperature ranges (0, 10, 28, 35 and 40⁰C) and NaCl concentrations (1-20%) on TSB medium.
Salt (NaCl) tolerance test
The NaCl tolerance was optimized for all the 23 isolates and fined their optimum growth on different NaCl percentage. Nine of the bacterial isolates grow best at salt concentration 0% , one of the bacterial strain (MMBK-DL) grow best at NaCl concentration 1%. Six of the bacterial strains grow best at NaCl concentration 2%, 5 of the isolates grow best at NaCl concentration 3% and two of the isolates grow best at concentration 10%. Growth ranges of the 17 isolates were 0-13%, two of the isolates growth range was 0-11% and five of the isolates have their growth range 0-17% salt concentrations as shown in the (Figure S1).
pH optimization test
All the 23 bacterial isolates were optimized to pH in the range of pH from acidic to basic. The optimum pH and their growth range of all the isolates are shown in the Figure 2. All the isolates showed maximum growth at neutral pH 7-9. None of the isolate could grow at pH 4 as well as at pH 10 (Figure S2).
Temperature optimization
No growth was seen at 0ºC, 4ºC, 20ºC. Maximum growth was observed at 35ºC and moderate growth was observed at 28ºC and 40ºC.
Human and animals are the two most important sources for the antibiotic-resistant organism’s entry into water environments. The organisms which carry antibiotic-resistance genes, finally find their way to be the part of mobile genetic elements (plasmids, transposons, integrons), from where it to spread among water and soil bacterial communities.14,15 In the present study, the pH of the water increased with increasing the distance from the source. This may be due to the gradual increase in a dissolved organic matter that increased the OD, electric conductivity, pH of the river water. Our results are in line with,16 who worked on River bank filtration in Haridwar, India. Most of the isolates were gram-negative rods. These results are in agreement with the study of.17 We found that culture-able antibiotic-resistance bacteria were present there in Swat River. About (95.45%) bacterial isolates were found resistant to Ampicillin, while the least percentage of the isolates (31.81%) showed resistance to Ciprofloxacin. Similar results were obtained by,18–21 where they isolated bacteria from the tap and fecally contaminated water, which showed resistance to one or more antibiotics. This level of isolates showed resistance to antibiotics because different Pharmaceuticals companies’ wastes are directly entered to the river without proper treatments and on the bank of the river there are different types of orchids for which farmers use the pesticides, during runoff they may enter into the river water. Recently an antibiotic resistance Pseudomonas spp was isolated from River Danube.21 The API strip results of identified species of the isolated bacteria are shown in Table 3. The API strip was used to identify different types of bacteria isolated from River Rouge.22 Most of the genera through API identification were pathogenic for humans like Pseudomonas spp, Proteus spp, Vibrio spp, Aeromonasspp, Salmonella spp, Serratiaspp,and Providencia spp. Different studies have been reported bacterial resistance genes and those bacteria which can be potential human pathogens, especially those which can cause gastrointestinal infections.23–26 Through API 20E strip we confirmed that most of the isolates were belong to genera Pseudomonas some studies reported Pseudomonas spp., from the fresh water.27
Codes |
API20E reagents |
Species identified |
|||||||||||||||||||
|
ONPG |
ADH |
LCD |
ODC |
CIT |
H2S |
URE |
TDA |
IND |
VP |
GEL |
GLU |
MAN |
INO |
SOR |
RHA |
SAC |
MEL |
AMY |
ARA |
|
MMBK.Mb |
- |
+ |
+ |
+ |
- |
- |
- |
- |
- |
- |
+ |
- |
- |
- |
- |
- |
- |
- |
- |
- |
Aeromonassalmonicidasspsalmonicida |
MMBK-Ma |
- |
+ |
- |
- |
+ |
- |
- |
+ |
- |
+ |
+ |
- |
- |
- |
- |
- |
- |
- |
- |
- |
Pseudomonas fluorescens/putida |
MMF1a |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
- |
- |
+ |
+ |
- |
- |
- |
- |
- |
- |
- |
- |
Proteus mirabilis |
MMC3e |
- |
- |
+ |
- |
+ |
- |
- |
+ |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
Stenotrophomonasmaltophilia |
MMBK-Ma |
- |
- |
+ |
+ |
+ |
- |
+ |
- |
- |
- |
+ |
- |
+ |
- |
- |
- |
- |
- |
- |
- |
Vibrio parahaemolyticus |
MMKC |
- |
+ |
- |
+ |
+ |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
- |
+ |
- |
+ |
- |
+ |
- |
Aeromonashydrophila/caviae/sobria 2 |
MMBK-Sm |
- |
+ |
+ |
+ |
+ |
- |
- |
- |
- |
- |
- |
+ |
- |
- |
- |
- |
- |
+ |
- |
+ |
Salmonella spp/ |
Pseudomonas luteola |
|||||||||||||||||||||
MMC2a |
- |
+ |
- |
- |
+ |
+ |
+ |
- |
- |
- |
+ |
+ |
- |
- |
- |
- |
+ |
- |
- |
- |
Proteus vulgaris group |
MMF1b3 |
+ |
- |
- |
- |
+ |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
- |
+ |
- |
+ |
- |
Serratiamarcescens |
MMF1b4 |
- |
+ |
+ |
- |
+ |
- |
+ |
- |
- |
+ |
+ |
- |
+ |
- |
- |
- |
+ |
- |
+ |
- |
Aeromonashydrophila/caviae/sobria 2 |
MMBK-DL |
+ |
+ |
+ |
+ |
+ |
+ |
- |
- |
- |
- |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
Salmonella spp |
MMBK-Dk |
- |
+ |
- |
- |
+ |
- |
- |
+ |
- |
+ |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
Pseudomonas fluorescens/putida |
MMBK-Dy |
- |
- |
- |
- |
- |
- |
+ |
+ |
+ |
- |
- |
- |
+ |
+ |
- |
- |
- |
- |
- |
- |
Providenciaretegeri |
MMF1b5 |
+ |
+ |
- |
- |
+ |
- |
+ |
- |
- |
+ |
+ |
- |
- |
- |
- |
- |
- |
- |
- |
- |
Pseudomonas aeruginosa |
MMBK-Sf |
Serratiaodorifera 1 |
||||||||||||||||||||
+ |
- |
- |
+ |
- |
+ |
+ |
+ |
+ |
- |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
||
MMBK-Sd |
- |
- |
+ |
- |
+ |
- |
+ |
+ |
+ |
- |
+ |
+ |
+ |
+ |
- |
+ |
- |
- |
- |
- |
Providenciarettgeri |
MMF1a1 |
- |
+ |
+ |
- |
+ |
- |
- |
- |
- |
+ |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
Pseudomonas putida |
Table 3 Analytical profile index (API 20E) of the bacterial isolates of river swat
The optimum pH, salt concentration and temperature of the current isolates showed that these microbes were adapted to 1% salt concentration and have a wide range of pH and temperature. This adaptation or tolerance in the isolates was because of the physical parameters of the river's water fluctuates during the entire year. Our results are similar to the previous studies.28–30 They isolated bacteria which grow best at the range of 0-1% salt concentration from Huangpu River-Yangtze River estuary. Four isolates were sequenced for 16S rRNA and all the isolates showed to be the members of Bacillus genera. Several researchers like31,32 and22 also used 16S rRNA for phylogenetic analysis of bacteria isolates from environmental samples. From the few isolates, we cannot assume that the diversity of bacteria is less in the river Swat but this is because of selective isolates. Isolate MMF1b2 showed similarity with Bacillus velezensis33 and this bacterial isolate play role in plant growth promotion.34 The other bacterial isolate, MMBK-M2a is closely related to Bacillus cereus have been reported by different studies35,36 its role in heavy metals remediation. MMF3a showed its sequence similarity with Bacillus subtilis, which now a day used as gram-positive bacteria model37 and also can produce very important hydrolytic enzymes of biotechnological importance.38–42 The bacterial isolate MMBK-Sc showed similarity with Bacillus axarquiensis, which previously have been isolated from river Vélez in Málaga, southern Spain.43 Bacillus axarquiensis can grow in a wide range of temperature and tolerate salt concentration. It can produce enzymes such as lipase, protease, and amylase. This bacterial isolate can be used for potential biotechnological purposes such as in leather and detergent industry.44
River Swat has a variety of bacteria, including isolates that are pathogenic to human. The presence of antibiotic-resistant isolates of bacteria in the river water may cause water-borne diseases in those areas. Along with the antibiotic resistance bacterial found in the river Swat, there was a positive aspect of the isolates was also observed. Some of the bacterial isolates from the river swat has great biotechnological applications like in remediation of heavy metals pollution, detergent and leather industries. The 16S rRNA gene sequencing analysis showed the phylogenetic relationship among the bacterial isolates of river Swat at different sites. Overall this study showed that River Swat can be explored further for novel and market value potential bacteria with addition of using biochemical and Sanger sequencing to explore the microbial potential for biotechnological applications and with health risk related to river water of Swat, Pakistan. The antibiotics resistance in microbes is consider one of the natural pollutants which are of prime importance for the fresh water, foods from rivers (fish) and the using of this water for irrigation purposes. On the other hand the research work also pointed out the rich sources of geoscience (hydrology) useful products from the natural sources like microbes of water. We suggest that proper treatment for the pharmaceutical industry is of prime importance before it disposes of off to the environments like rivers.
The authors are thankful to the Department of Microbiology, Kohat University of Science and Technology, Kohat, Pakistan for their support and help to fulfill the work.
All the authors have declared no conflict of interests.

©2021 Afzal, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.