Journal of
eISSN: 2469 - 2786


Research Article Volume 9 Issue 3
1Zhejiang University, China
2Department of Microbiology, Quaid-i-Azam University, Pakistan
3Islamic girls public school and college, Pakistan
4Department of Biotechnology, Bacha Khan University, Pakistan
5Department of Microbiology, Kohat University of Science and Technology, Pakistan
Correspondence: Muhammad Afzal, Zhejiang Provincial Key Laboratory of Subtropic Soil and Plant Nutrition, College of Environmental and Resource Sciences, Zhejiang University, Hangzhou, 310058, P.R. China
Received: July 03, 2021 | Published: July 14, 2021
Citation: Afzal M, Ihsan-Ud-din, Rafiq M, et al. Bacteria isolated from a limestone cave and its ability to produce industrially important enzymes. J Bacteriol Mycol Open Access. 2021;9(3):104-113. DOI: 10.15406/jbmoa.2021.09.00305
Caves have extreme environment with diverse microbial flora. In the present study a bacterial isolates, KC5- MRL, AF-3, AF-6 and AF-23 isolated from Kashmir Smast(cave), a limestone cave in PirsaiMardan,, KPK, Pakistan, was found to produce three industrially important enzymes; lipase, protease and amylase. The bacterial isolates were identified as Lysinibacillus sphaericus KC5-MRL, Bacillus subtilis AF-3, Bacillus subtilis AF-6 and Bacillus subtilis AF-23(Accession No. KF010827, MH027600, MH027601 and MH027602), respectively. Optimum pH for the growth of Lysinibacillus sphaericus KC5- MRL, was around 7 and grew best at 35ºC. The optimum activities of lipase and amylase was observed at 30°C after 24 hr of incubation and pH 5 (42.23 U/ml) and (15U/ml) respectively. Maximum lypolytic activity (181.93 U/ml)was observed when 8% inoculum was used. Proteolytic activity of L. sphaericus KC5-MRL was found to be 59 U/ml, after 48 hr at 30°C.Highest stability of lipase, amylase and protease was observed at pH 10, 7 and 8 respectively. L. sphaericus KC5-MRL lipase, protease and amylase were stable at 35ºC.Triton X-100 and sodium dodecyl sulfate (SDS) stimulated the lipase and protease activities, whereas, Triton X-100 and T-80 stimulated amylolytic activity. Lipase activity was stimulated by Mg++, NH4+ and Ca++. Amylase activity was stimulated by Na+ and Ca++.Protease activity was stimulated by Na+ and Mg+. Chloroform, formaldehyde, methanol and benzene stimulated amylase activity. Nitrobenzene, methanol, benzene, and acetone stimulated protease activity. Ethylenediaminetetraacetic acid (EDTA) showed stimulatory effect on lipase. EDTA, Trisodium citrate TSC, mercaptoethanol, Phenyl acetaldehyde (PAA) and PMSF reduced amylase activity. All the modulators reduced protease activity except TSC and PMSF which stimulated its activity. The study concludes that these enzymes can be used for different purposes in various industries such as food and detergent.
Keywords: KC5- MRL, Kashmir smast, enzymes, limestone, Pakistan
Caves are known to have very poor nutrient ecology and have constant low temperature and high humidity. Because of this environment, the cave organisms have some extraordinary properties to survive and live in such hard and crucial condition.1–3 Cave ecology is rich in different types of microorganisms having outstanding characteristics. The most abundant organisms observed in caves are filamentous and belong to Actinobacteria group, followed by coccoid and bacilli forms.4 A few pathogenic microorganisms have been reported from the Altamira cave.5 Luonget al.6 described for the first time, A. altamirensis recovered from human medical constituents. E. coli and S. aureus which are disease causing bacteria have also been isolated from caves,7 species of Pseudomonas, Sphingomonas and Alcaligenes sp8 and Inquilinusp.9 Nutrient recycling in the cave totally depends upon the bacterial and fungal activities.10 Caves have fewer amounts of nutrients, less light or no light ecosystem, which have relatively low temperature and high humidity. From different caves, around the world, different types of microbial species have been isolated.11 Most of the caves which have been studied are in Spain, Italy, France, Romania and USA. Altamira Cave in Spain and Lascaux Cave of France is the most microbiologically studied cave.12,13 Significant growth in biospeleological research has been observed in the last two decades.14 Caves are considered as extreme environments providing niches for highly specialized microorganisms.15 Enzymes which are isolated from cave microbes are active at low temperature. Enzymes of psychrotrophs have great biotechnological importance.11 These microbes survive in such environment because of production of enzymes which function in such extreme conditions.16 Our knowledge about cave microbial diversity is limited, so there is great potential to find novel microorganisms in caves.17 Caves in Pakistan are less studied area especially with regards to industrially important enzymes from cave bacteria have not been reported or isolated so for. Therefore, in our study we focused, on the isolation of bacteria from Kashmir cave, and their potential to produce industrially important enzymes having some unique properties.
The present study was carried out in the Microbiology Research Laboratory (MRL), Department of Microbiology, Quaid-i-Azam University, Islamabad, Pakistan. Production of lipase, amylase and protease by bacterial isolates, isolated from a limestone cave (Kashmir Smast Pirsai Mardan, Pakistan) (Figure 1) was investigated. The cave (GPS co-ordinates 34° 25' 48.24" N 72° 13' 43.18" E) is 188m long, 30m high and 25m wide, is located in Pirsai Mardan, Pakistan. Soil samples were collected in sterile bags under aseptic conditions.
Protease, Lipase and Amylase
Nutrient agar plates containing (1%) casein as substrate were used to check the proteolytic activity which was observed in the form of clear zones of hydrolysis around the colonies. For the screening of lipase, Tween-80 was used as lipid substrate. After incubation at 37°C for 24h, appearance of clear zones was observed. Starch was used as substrate to detect the production of amylolytic enzyme. Inoculated plates were incubated at 37°C for 24h and clear zones of hydrolysis were observed.
PCR amplification and sequencing of 16S rRNA gene
To amplify the 16S rRNA gene the genomic DNA was extracted according to Roohiet al. (2012). Briefly, few isolated bacterial colonies were suspended in TE buffer in a micro-centrifuge tube. Followed by heating for 10 min at 95ºC and centrifugation at 6,000 rpm for 5 min. to amplify the 16S rRNA gene the supernatant was used as template DNA. Polymerase chain reaction (PCR) was used to amplify the 16S rRNA gene, using primers 9F (5'-GAGTTTGATCCTGGCTCAG-3') and 1510R (5'-GGCTACCTTGTTACGA-3') using Premix Ex Taq (Takara, Japan) following protocol described previously. The amplification was carried out under the optimized PCR conditions: initial denaturation at 94ºC for 2 min; 30 cycles of denaturation at 94ºC for 1 min, annealing at 50ºC for 1 min and extension at 72ºC for 1:30 min. The final extension was performed at 72ºC for 5 min in ABI Veriti PCR Machine (Applied Biosystems, USA). The amplified PCR products of 16S rRNA gene of bacterial strain was purified and sequenced using the primers 27F (5' AGAGTTTGATCMTGGCTCAG-3') and 1492R (5'-ACCTTGTTACGACTT-3') using commercial service of Macrogen Inc. Korea (http://dna.macrogen.com/eng/).
Phylogenetic analysis of bacterial isolates
BioEdit software (Hall, 1999) was used to assemble the fragment sequences of 16S rRNA gene. The 16S rRNA gene sequences were submitted to NCBI (https://www.ncbi.nlm.nih.gov/). Using 16S rRNA gene sequences, the strain was identified by BLAST search on EzBiocloud Server). Closely related type species sequences were retrieved to assess the molecular evolutionary relations and to construct phylogenetic tree using MEGA version.6 A phylogenetic tree was constructed from unambiguously aligned nucleotides using the neighbour-joining (NJ) algorithm. The relationship stability was evaluated by boot strap analysis performed using 1000 re-samplings of the neighbour-joining data for the tree topology.
Quantitative analysis of lipase
About 150ml of medium was added in four different Erlenmeyer flasks, autoclaved for 20 minutes at 121°C and 15lbs. About 1.5ml of olive oil was added to the above medium which was already sterilized in oven at 121°C for half an hour. Then medium was inoculated with two loops full of culture under sterilized conditions, and then flasks were kept in shaking incubator at 37°C and 150rpm. Samples were drawn after 0, 24, 48 and 72h. Enzyme assay for lipase was performed by the method of.18 Unit of enzyme is defined as the amount of enzyme that hydrolyzes 1μmol substrate in 1 minute.
Quantitative test for protease
Medium (Gelatin: 15.0g/250ml, Casein hydrolysate: 0.5g/250ml, Glycerol: 3.0ml and pH 8) of 250ml was poured to four Erlenmeyer flasks, pH was adjusted by using 0.1N NaOH and 0.1N CH3COOH.The medium was autoclaved at 121°C 15lbs pressure for 20 minutes 3.0ml of 20% glycerol solution sterilized in dry oven at 100°C for half an hour was poured into the medium aseptically. Then two loops full of bacterial culture were inoculated in different flasks. Then flasks were kept in shaking incubator at 37°Cwith 150rpm. Samples were collected after 0, 24, 48 and 72hrs for quantitative analysis.
Enzyme assay for protease
Enzyme assay for protease (Kunitz,1965), briefly 1ml of casein solution (1% casein solution in 0.02M Tris buffer, pH 8.5) was added into 1ml of crude enzyme. The mixture was incubated at 45°C for 1hour.After incubation 3ml of 5% TCA was added. Then tubes were placed in ice for 15minutes the precipitates were removed by centrifugation at 5000rpm for 15 minutes at 4°C. About 5% TCA was added in blanks before incubation. The optical density of the supernatant was measured at 280nm. Graph for standard curve was prepared using tyrosine as standard. One unit of enzyme caseinolytic activity defined as that amount of enzyme which releases 1μ mol tyrosine under standard conditions of assay method.
Shake flask fermentation for optimization of different parameters
To determine the optimum pH for lipase production, fermentation was carried out at different pH (3, 4, 5, 6, 7, 8 and 9) and optimum temperature was determined by carrying out shake flask fermentation at various temperatures (15, 25, 30, 35 and 40°C). For the size of inoculums a 24h old bacterial culture grown in nutrient broth was added at a concentration of 1to10% to the production medium. About 150 ml of production medium was poured into four different flasks. Samples were taken after 0, 24, 48 and 72hr and assayed for lipase activity, and centrifuged at 10,000rpm for 30 minutes at 4°C. Precipitates were removed by filtration and supernatant was used as the crude enzyme for the estimation of enzyme activity.
Effect of temperature, pH, metals ions, modulators and solvents on enzyme activity
Activity of crude enzyme was determined after incubating for 1hr at 30, 35 40, 50, 60, 70 and 80 and 100˚C.Effect of pH on the activity of crude enzyme extract was studied at different pH. The crude enzyme was incubated for 1h in buffers having different pH [0.02 M of acetate buffer (pH 4, 5, 6), phosphate buffer (pH 6, 7), Tris-HCl (pH 8), glycine-NaOH buffer (pH 9) and Na2HPO4/NaOH (pH 10)], and remaining enzyme activity was determined under standard conditions. Effect of different metals (100mM of HgCl2, NaCl, MgCl2, ZnCl2, (NH4)2SO4 and CaCl2separately) on activity of enzymes. These metals were pre-incubated with crude extract (1:1) for 1hr at room temperature and then remaining activity was determined. Effect of different modulators on enzyme activity was determined for EDTA, Trisodium citrate, mercaptoethanol and PMSF. About 1% of these compounds were pre-incubated for 1h at room temperature (25°C) with crude enzyme (1:1). Effect of different solvents on activity of lipase was determined after pre-incubating crude enzyme in the presence of (1% each, separately) xylene, benzene, chloroform, nitrobenzene, formaldehyde, methanol and acetone (1:1) for 1h at room temperature. For protein estimation, Lowry’s method19 was used.
Statistical analysis
All statistical analyses were performed using SPSS version 20, and one-way analysis of variance (ANOVA) followed by LSD was used to check for the differences of enzymes activity under different treatments. P≤0.05 and P≤0.01 were statistically significant. The figures of differences of enzymes activity between treatments was prepared by Origin (Origin Pro 9.0 for Windows).
Primary screening for the enzymes (lipase, amylase and protease)
All the strains were Gram positive rods. Phylogenetic analysis shows that AF-3, AF-6 and AF-23 closely related to Bacillus subtilis and KC5-MRL was closely related to Lysinobacillus sparicous. On the basis of quantitative analysis Bacillus subtilis AF-6, Bacillus subtilis AF-3, Lysinobacillus sparicous KC5-MRL, and Bacillus subtilis AF-23 strains were positive for the enzymes and selected for further study. Growth of all the strains were optimum at neutral pH 7, at 35°C and almost no growth at temperature 15 and 40°C, and size of inoculum8% after 72h.
Effect of pH, temperature and size of inoculum for the optimum lipase, amylase and protease activity
The optimum activity of L.sphaericus KC5-MRL lipase was observed at 30°C after 24 hr of incubation and pH 5 i.e. 42.23U/ml. Maximum lypolytic (181.93U/ml), proteolytic (59 U/ml) and amylolytic (15U/ml) activity was observed when 7% of inoculum was used at 30°C and 24 h of incubation (Fig.). Bacillus subtilis AF-3 produced maximum lipase (20U/ml), amylase (9.74U/ml) and protease (40.5U/ml) at 6% inculum 24 and 72h of incubation at 35°C, 30°C respectively. Bacillus subtilis AF-6 produced maximum lypolytic (21U/ml), proteolytic (40.7U/ml) and amylolytic (15.59U/ml) at 6% of inculum after 24, 48 and 72h of incubation at 30°C, and pH 5, respectively. Bacillus subtilis AF-23 produced maximum lypolytic (18U/ml), proteolytic (77.5U/ml) and amylolytic (9.78U/ml) at 5% inculum after 48h of incubation at 25°C and 30°C and pH 6, respectively.
Characterization of crude lipase, amylase and protease produced by the isolates
Stability of crude enzymes at different pH: Amylolytic activity of sparicous KC5-MRL, B. subtilis AF-3, B. subtilis AF-6 and B. subtilis AF-23 showed highest stability at pH 7, 10, 8 and 10 i.e 99.4%, 101%, 99.89% and 107%, respectively. Proteolytic activity of L. sparicous KC5-MRL, B. subtilis AF-3, B. subtilisAF-6 and B. subtilis AF-23 were more stable at pH 8, 7, 8 and 10 i.e 115%, 127%, 106% and 102 respectively. Proteases of all the four strains were stable at all pH values. Lipases of all the strains were not stable at pH 4. Highest stability of L. sparicous KC5-MRL, B. subtilis AF-3, B. subtilis AF-6 and B. subtilis AF-23 lipases were observed at pH 10, 7,8 and 6i.e. 42%, 40.5%, 34.5% and 41% respectively.
Stability of crude enzymes at different temperature: Lowest satiability of lipase of all the isolates was observed at 100°C. sphaericus KC5-MRL, B. subtilis AF-3, B. subtilis AF-6 and B. subtilis AF-23 lipases highest stability were observed at 35°C (118%), 40°C (80%), 30°C(105%) and 40°C (40%), respectively. The highest temperature stability of L. sphaericus KC5-MRL, B. subtilis AF-3, B. subtilis AF-6 and B. subtilis AF-23, amylases were observed at 35°C (107%), 70ºC( 107%), 30°C (102%) and 40°C (122%). Lowest stability of amylases were noted at 100C (0%), 50ºC (94%), 100C (14%) and 50C (33%) for L. sphaericus KC5-MRL, B. subtilis AF-3, B. subtilis AF-6 and B. subtilis AF-23, respectively. The highest temperature stability of proteases of L. sphaericus KC5-MRL, B. subtilis AF-3, B. subtilis AF-6 and B. subtilis AF-23, were observed at 35°C(104%), 80ºC (127%), 40°C (149%) and 100°C (111%), respectively. Lowest temperature stability was observed at 100C (31%), 60ºC (25%), 70C (55%) and 50C (83%) for L. sphaericus KC5-MRL, B. subtilis AF-3, B. subtilis AF-6 and B. subtilis AF-23, respectively.
Effect of detergents on enzymes activity
Addition of Tween-80 reduced liypolytic activity of L. sparicous KC5-MRLup to 4%. Triton X-100 (T X-100) and sodium dodecyl sulfate (SDS) stimulated the lipase activity up to 4% and 3%, respectively. Triton X-100, Tween-80 (T-80) and SDS reduced the B. subtilis AF-3 lipase activity up to 4%, 6% and 9% respectively. While reduced the activity up to 4%. While Triton X-100 stimulated the lipase activity up to 6%. Tween-80 and SDS stimulated the B. subtilis AF-6 lipase activity up to 5% and 7% respectively. While Triton X-100 reduced the activity up to 2%. Triton X-100 and SDS stimulated the B. subtilis AF-23 lipase activity up to 5% and 4% respectively. While Tween-80, reduced lipase activity up to 2%.SDS reduced amylase activity of L. sparicous KC5-MRL upto 70%. TX-100 and T-80 stimulate enzyme activity 19 and 23% respectively. SDS completely inhibited amylase of B. subtilis AF-3 enzyme activity. TX-100 and T-80 showed inhibitory effect inhibited activity upto 98 and 19%.SDS reduced B. subtilis AF-6enzyme activity upto 45.5%. TX-100 and T-80 stimulate enzyme activity 5 and 8.5% respectively. SDS and TX-100 reduced B. subtilis AF-23enzyme activity upto 4.3 and 1% and T-80 stimulate enzyme activity up to 9%.T-X-100 and T-80 showed stimulatory effect, stimulated L. sparicous KC5-MRL protease activity upto 9 and 23% respectively. SDS reduced lipase activity up to 70%. Protease of B. subtilis AF-3 stimulated by TX-100 upto 82%. SDS and T-80 reduced protease activity of B. subtilis AF-6 up to 31 and 4%, respectively. Triton X-100 and SDS showed stimulatory effect, stimulated B. subtilis AF-6 protease activity upto 48.7 and 28%, respectively while T-80 reduced enzyme activity 62.3%.TX-100 and T-80 showed stimulatory effect, stimulated B. subtilis AF-23 protease activity upto16% and 31%, respectively, while SDS reduces enzyme activity up to 66.5%.
Effect of metal ions on enzymes activity: All the metals were inhibitory for all proteases and amylases of subtilis AF-3 and B. subtilis AF-6. For the rest of enzymes the some metals showed stimulatory and some inhibitory effect. The metal ions e.g. Mg++, NH4+ and Ca++stimulated lipase activity of L. sparicous KC5-MRL as; 25.8%, 31.1% and 27.54%, respectively. Ammonium ion stimulated the lipase activity up to 31.7%. Hg+Na+ and Zn++ inhibited lipase activity as 2%, 7% and 11.31%, respectively. Zn++ showed highest inhibitory effect on lipase activity. The metal ions e.g. Na+, Mg++and Ca++ stimulated lipase activity of B. subtilis AF-3 up to 20%, 27% and 31%, respectively. Ammonium ion, Hg+ and Zn++ inhibited lipase activity as 6%, 10% and 18%, respectively. Zn++ showed highest inhibitory effect on lipase activity. The metal ions e.g. Mg++and Ca++ stimulated lipase activity of B. subtilisAF-6up to 26% and 30%, respectively. Hg+, Na+ and Zn++ inhibited lipase activity as 4%, 8% and 12%, respectively. Ammonium ion and Zn++ showed highest inhibitory effect on lipase activity. The metal ions e.g. Mg++and Ca++ stimulated lipase activity of B. subtilis AF-23 as; 27% and 31%, respectively. Ammonium ion, Hg+, Na+ and Zn++ inhibited lipase activity as 3%, 6%, 10% and 23%, respectively. Zn++ showed highest inhibitory effect on lipase activity. Some metals were inhibitory and some stimulatory for amylase enzyme. L. sparicous KC5-MRL amylase showed reduced activity under Hg++, Mg++, Zn++ and NH4+ treatment as; 69%, 11%, 26% and 55% respectively. Na+ and Ca++ showed stimulatory effect up to 11 and 6% respectively. Metals ions showed inhibitory effect on amylase activity of B. subtilis AF-3 crude extracts. Ca++ completely inhibits the enzyme activity. Hg++, Na+, Mg++ reduced enzyme activity up to 33, 54 and 25% respectively. B. subtilis AF-6amylase activity was reduced by Hg++, Na+, Mg++, Zn++, NH4+and Ca++up to 72%, 12%, 2%, 28%, 18% and 21% respectively. B. subtilis AF-23 amylase activity was reduced by Hg++, Na+, Mg++, Zn++and NH4+ up to 79%, 44%, 100%, 81.5% and 75.5% respectively while Ca++showed stimulatory effect up to 84%.All metals were inhibitory for B. subtilis AF-3 protease enzyme. Hg++, Na+, Mg++, Zn++, Ca++and NH4+reduced enzyme activity up to 17%, 49%, 74%, 64%, and 64% respectively. All metals were inhibitory for B. subtilis AF-6protease. Hg+, Na+, Mg++, Zn++, Ca++and NH4+reduced enzyme activity up to 72%, 12%, 1.4%, 72%, 21% and 18%, respectively. Some metals were inhibitory for L. sparicous KC5-MRLprotease. Hg++, Zn++, Ca++and NH4+reduced enzyme activity up to 57%, 46%, 47% and 15%, respectively. Na+ and Mg++stimulate L. sparicous KC5-MRL protease activity up to 8% and 16% respectively. All metals were inhibitory for B. subtilis AF-23protease except Na+, which stimulated enzyme up to 65%. Hg++, Mg++, Zn++, Ca++and NH4+reduced B. subtilis AF-23 protease activity 43%, 50%, 23.4%, 60.3% and 38.2%, respectively.
Effect of organic solvents on enzymes activity: Presence of nitro-benzene (N.B) reduced lipase activity of sparicous KC5-MRLup to 3.4%. Other organic solvents stimulated lipase activity. Highest stimulation was observed in case of benzene (B) 14.32%. Addition of xylene (X) showed no effect on the activity of lipase while chloroform (Ch), formaldehyde (Frm), methanol (Met) and acetone (Ace), stimulated lipase activity as 2.5%, 10.5%, 4.2% and 3%, respectively. Presence of N.B reduced lipase activity of B. subtilis AF-3 up to 5%. Other organic solvents stimulated lipase activity. Highest stimulation was observed in case of Ace16%. Addition of X and methanol showed no effect on the activity of lipase while Ch, Frm and B stimulated lipase activity as 3%, 9%, 5% and 4%, respectively. Presence of N.B reduced lipase activity of B. subtilisAF-6up to 8%. Other organic solvents stimulated lipase activity. Highest stimulation was observed in case of Ace 11%. Addition of X and methanol showed no effect on the activity of lipase while Ch, Frm and benzene stimulated lipase activity as 2%, 4%, 6% and 3%, respectively. Presence of N. B and Ch reduced lipase activity of B. subtilisAF-23up to 6% and 8%. Other organic solvents stimulated lipase activity. Highest stimulation was observed in case of Ace13%. Addition of X and methanol showed no effect on the activity of lipase while Frm and B stimulated lipase activity as 7% and 4%, respectively. Nitrobenzene X and Ace reduced amylase activity of L. sparicous KC5-MRL up to 33%, 5% and 50%, respectively. Chloroform, Frm, Met and B addition stimulated L. sparicous KC5-MRL amylase activity upto 9%, 4%, 3.5%, 2% and 7%, respectively. Benzene and Ch reduced B. subtilis AF-3 amylase enzyme activity up to 18 and 90% respectively. Nitro Benzene stimulation was highest 86%, X, Frm, Ace and Met stimulate enzyme activity up to 70%, 60%,56% and 48%, respectively. All organic solvents reduced B. subtilis AF-6 enzyme activity. Chloroform completely inhibited enzyme activity. Nitro Benzene, X, Frm, Ace, Met and B reduced B. subtilis AF-6 enzyme activity up to 16%, 19%, 13%, 15% and 17%,respectively.. Chloroform, Frm, Met, B, X and Acereduced B. subtilis AF-23amylaseactivity up to 87.5%, 34%, 66%, 32% and 63%, respectively. Nitrobenzene, Met, B, and Ace stimulated L. sparicous KC5-MRL protease activity upto17%, 17%, 4% and 27%, respectively. Formaldehyde, Ch and X reduced the activity of protease from L. sparicous KC5-MRL up to 10%, 66% and 30%, respectively. Chloroform, Frm, Met, B, X and Acereduced B. subtilis A F-3protease activity up to 17%, 100%, 77%, 33%, 45% and 74.5%, respectively. Nitrobenzene, Frm, B, and Acestimulate B. subtilisAF-6 enzyme activity up to 22%, 1.5%, 37% and 20.5%, respectively. Methanol, Ch and X reduced the activity of protease from B. subtilis AF-6 up to 5%, 63.4% and 34.5%, respectively. Chloroform showed no effect on the activity on B. subtilis AF-6 protease. Nitrobenzene, X and Frm reduced B. subtilis AF-23proteaseactivity up to 42%, 66.5% and 30.4%, respectively. Acetone and Met stimulated B. subtilis AF-23proteaseactivity up to15.5% and 30.4% respectively.
Effect of modulators on enzymes activity: Phenyl acetaldehyde (PAA) inhibited the activity of crude extract of lipase of Sparicous KC5-MRLup to 17.3%. Ethylenediamine Tetraacetic acid (EDTA) stimulated lipolytic activity up to 24.4%. Mercaptoethanol (Mre) showed no effect on lipolytic activity, whereas, Tri-sodium citrate (TSC) and phenylmethylsulfonyl fluoride (PMSF) both inhibited lipase activity up to 14%. Phenyl acetaldehyde inhibited the activity of crude extract of lipase of B. subtilis AF-3 up to 18%. EthylenediamineTetraacetic acid stimulated lipolytic activity up to 24%. Mercaptoethanol showed no effect on lipolytic activity, whereas, TSC and PMSF both inhibited lipase activity up to 11% and 13%, respectively. Phenyl acetaldehyde inhibited the activity of crude extract of lipase of B. subtilis AF-6 up to 18%. Ethylenediamine Tetraacetic acid stimulated lipolytic activity up to 23%. Mercaptoethanol showed no effect on lipolytic activity, whereas, TSC and PMSF both inhibited lipase activity up to 15%. Phenyl acetaldehyde inhibited the activity of crude extract of lipase of B. subtilis AF-23 up to 20%. Ethylenediamine Tetraacetic acid stimulated lipolytic activity up to 21%. Mercaptoethanol showed no effect on lipolytic activity, whereas, TSC and PMSF both inhibited lipase activity up to 18% and 23%, respectively. All the inhibitors reduced the amylases activity of all the strains except for amylase of B. subtilis AF-3 which was stimulated by PAA up to 43%. L. sparicous KC5-MRL amylase activity inhibited by EDTA, TSC, Mer, PAA and PMSF up to 18%, 84%, 01%, and 16%, respectively. The amylase B. subtilis AF-3activity inhibited by EDTA, TSC, Mer and PMSF up to 93%, 43%, 59%, and 51%, respectively. B. subtilis AF-6amylaseactivity was reduced by EDTA, TSC, Mer, PAA and PMSF up to 55.5%, 54%, 32%, 16% and 37%, respectively. EDTA, The amylase activity of B. subtilis AF-23 reduced by TSC, Mer, PAA and PMSF up to 01%, 01%, 79%, and 100% and 100%, respectively. All the inhibitors reduced the protease activity of L. sparicous KC5-MRL except TSC and PMSF which stimulated L. sparicous KC5-MRL protease a ctivity up to 24 and 5% respectively. Ethylenediamine Tetraacetic acid, Mer and PAA reduced L. sparicous KC5-MRL protease activity up to 34, 37, and 26%, respectively. All the inhibitors reduced B. subtilis AF-3 protease enzyme activity EDTA, TSC, Mer, PAA and PMSF reduced enzyme activity up to 64%, 66%, 26%, and 81% and 56%, respectively. All the inhibitors reduced B. subtilis AF-6 enzyme activity except PAA which stimulated enzyme activity up to 69%. Ethylenediamine Tetraacetic acid, TSC, Mer and PMSF reduced enzyme activity up to 61%, 36%, 25%, and 68%, respectively. All the inhibitors reduced B. subtilisAF-23 enzyme activity. Tri-sodium citrate, PMSF, EDTA and Mer reduced enzyme activity up to 46%, 50%, and 33% respectively.
Enzymes are known as nature’s catalysts.20 So for different enzymes are isolated from different living organisms like plants, animals, fungi, yeast and bacteria.21 Throughout the world only 2% enzymes are isolated from microorganisms.22 These microorganisms are isolated from different ecosystems. The hidden potential of microorganisms in sense of valuable industrial enzymes exploration is still on wait. These enzymes have number of applications in different industries like food, medicine, cosmetics, textile, leather, detergents, oleo chemical and for different diagnostic tools (Hasanet al., 2005). Caves are extreme environment with poor nutrient content. Caves still have to be explored for new horizons in Microspeleology sectors. In the world different antibiotics are isolated from different caves. From ice caves different types of industrially important enzymes have also been reported. Our present study is focused on caves bacteria to find out industrially important enzymes. Lysinibacillus sphaericus is a gram positive, rod shaped mesophilic bacterium that naturally occur in soil. Lysinibacillus sphaericus KC5-MRLshowed maximum lipase production 42.23U/ml at 30°C and pH 5, after 24h of incubation. Similar specific enzyme activity was reported by different investigators for lipase.23-25 Lipase from Pseudomonas sp., which produced maximum lipase after 24h of incubation at 30-40°C.26 Pseudomonas sp., produced maximum lipase at pH 5.5.27 Variation in pH, incubation time and incubation temperature may be due to isolate specificity for different conditions.
Maximum lipase activity was observed at a size of 7% inoculum. Balanet al.,28 reported maximum lipase production when 7% inoculum of Geobacillus thermodenitrifican was used in flask fermentation. Effect of different metal ions was checked on the basis of residual activity of crude extract. Mg++, NH4+ and Ca++ stimulated lipase activity. Stimulation of crude extract activity have been reported by different investigators Li and Ziaobo (2005) reported that Ca++ and Mg++ stimulate the activity of Geobacillussp.TW1 lipase. Our results are similar to that of Mobaraket al.29 where Pseudomonas aeruginosa KM110 lipase was inhibited by Zn++ and Cu++ .29 Organic solvents mostly showed some kind of stimulatory effects. Highest stimulation noted in case of benzene while nitrobenzene was inhibitory for enzyme. Our results are similar with that of Nawaniet al.30 They reported benzene as stimulator for the lipase of thermophilic Bacillus sp. Schmidtet al.31 reported that different organic solvents like methanol, acetone and ethanol enhanced lipase activity isolated from B.thermocatenulatus. Lee et al.32 reported that different detergents change the activity of Bacillus thermoleovorans ID-1thermophilic lipase. They reported no activity of Triton X-100 but in contrast to our study, it stimulated the lipase activity and SDS also stimulated enzyme activity. Bacillus thermoleovorans ID-1 lipase activity were reduced by Tween 80. Mobaraket al.29 reported that lipase of Pseudomonas aeruginosa KM110 showed optimum activity at pH8 i.e. 27%. Our isolated lipase also showed 142% activity at alkaline pH 10. This was maximum activity for the pH value tested. Kojima et al.,(1994) isolated bacterial lipase from Pseudomonas fluorescens AK102 which were stable at pH 4-10 and maximum stability was noted at pH 8-10. Lipase retained its activity at all temperatures tested. The highest activity was observed at 35°C. The enzymes were stable at 30-80°C. Wang et al.33 characterized a bacterial lipase of Bacillus strain A30-1 (ATCC 53841) which was stable at 75°C. Rathiet al.34 isolated a lipase from Pseudomonas sp., which was stable at 90°C for 3h. In our research, lipase was stable for 1 h at 80°C. Demirkan35 isolated amylase from Bacillus subtilis reported highest activity from the current study. Different investigators36,37 have reported that protease is produced during stationary phase. They isolated these proteases from Pseudomonas fluorescens P26 and Bacillus spp. Amroet al.38 isolated bacterial proteases from different bacteria which have maximum protease activity in the range of 34-44 U/ml. In our study we reported 59 U/mlproteases activity. Overall, the isolated amylase in our study were stable from 30 to 80ºC for 1h. Malhotraet al.39 reported α-amylase of Bacillus thermooleovorans NP54which was active at 100ºC for 10 minutes. The enzymes they reported were stable at 40-100ºC.
Our results are similar to that of Adinarayanaet al.40 They isolated protease from Bacillus sp. BZI-2 and Femi-Ola & Oladokun41 isolated protease from Lactobacillus brevis. Pathak & Deshmukh42 reported a protease from Bacillus licheniformis which was active in the range of 20-90ºC. Lee et al.43 isolated a protease from Pseudoalteromonas sp. A28, which is in accordance with our study. Some metals were found inhibitory and some stimulatory for the Lysinibacillus sphaericus KC5-MRLamylase. Hailemariamet al.44 also reported amylase from a Bacillus sp. which was inhibited by different metals ions. Activity of amylase from different bacterial isolates was reduced by different divalent cations45-50 reported two different types of bacterial amylases whose activities were inhibited by chloroform and other organic solvents while alcohol and acetone stimulated enzymes activity. Similar observation was reported in our study. All the inhibitors reduced the amylases activity. Ballschmiteret al.51 reported an amylase from hyper thermophile bacteria with reduced activity in the presence of EDTA. They reported that amylase activity was slightly stimulated by the presence of mercaptoethanol which contradicts with our results. None of the surfactants tested had a pronounced inhibitory effect on amylase activities. Oliveiraet al.52 reported amylase from Rhizobia strains. Amylase activity was not inhibited by detergents/ surfactants which they used in their experiments. They reported that Triton X-100 moderately inhibited enzyme activity and SDS and Tween-80 showed stimulatory effect on the enzyme activity. Similar results were noted in the present study. Addition of surfactants in case of proteases showed some stimulatory and as well as inhibitory effect. Nascimento &Martins53 isolated protease from Bacillus sp., which was inhibited by the addition of surfactants like SDS and Triton X-100. This result is similar to the present study.
All metals were inhibitory for the protease. Banerjeeet al.54 reported Brevibacillus (Bacillus) brevis to be inhibited by CuSO4, ZnCl2 and HgCl2. Usharaniand Muthuraj55 reported Bacillus laterosporus protease whose activity was inhibited by the presence of different metals ions. The inhibitory effect of heavy metal ions is well documented in the literature. In the present study most of the metal ions inhibited the enzyme activity. Organic solvents showed stimulatory and as well as inhibitory effects in case of proteases. Rahman et al.56 isolated protease from Pseudomonas aeruginosa strain K whose activity was inhibited by 1-pentanol, benzene, toluene,p-xylene and n-hexane up to 33%, 35%, 30%, 48% and 36% respectively. Protease activity was stimulated by metals ions. Lysinibacillus sphaericus KC5-MR showed that this protease belong to metalo proteases. Similar results also reported by Mabrouket al.57 they isolated protease from Bacillus licheniformis ATCC 21415, which was inhibited by EDTA. The hydrolytic activity of different enzymes (Lipase, protease, amylase) was characterized. Lysinibacillus sphaericus KC5-MRL has shown varying activities of enzymes. Lysinibacillus sphaericus KC5-MRL enzymes properties indicated possibilities to be used for enzyme biotechnology in order to enhance catalytic efficiency of enzymes in industry (Figure 2 – Figure 10).58–63

Figure 2 Growth of strain and lipase production of Lysinibacillus sphaericus KC5-MRL at different sizes of inoculum.
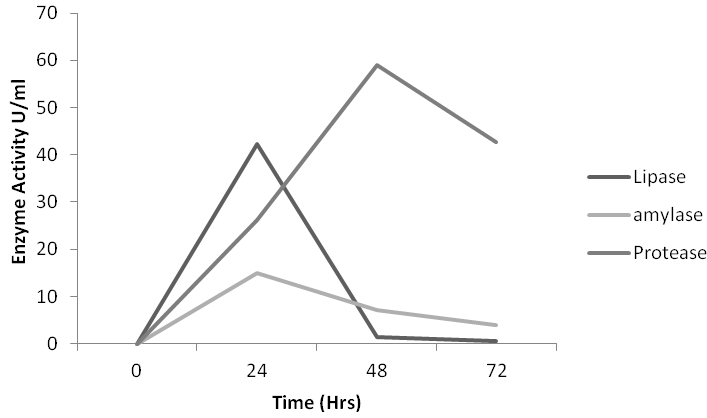
Figure 3 Lipolytic, Amylolytic and proteolytic activity of strain Lysinibacillus sphaericus KC5-MRL at 30°C.
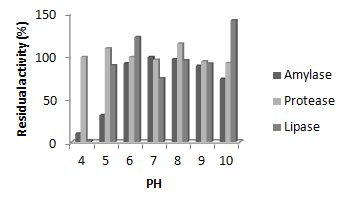
Figure 4 Stability of crude extract of lipase, amylase and protease of Lysinibacillus sphaericus KC5-MRL at different pH.
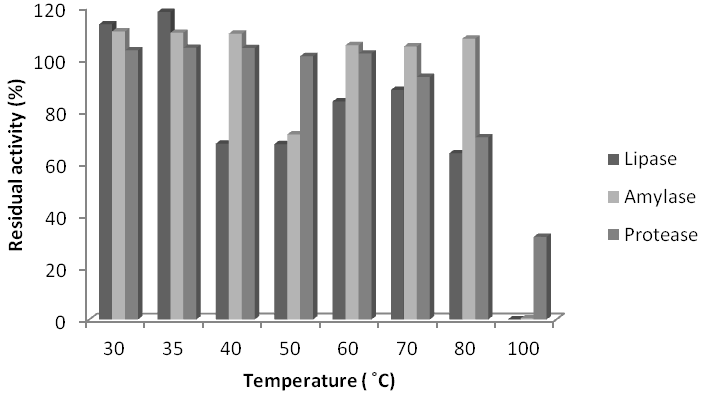
Figure 5 Stability of crude extracts of lipase, amylase and protease of Lysinibacillus sphaericus KC5-MRL at different temperature.
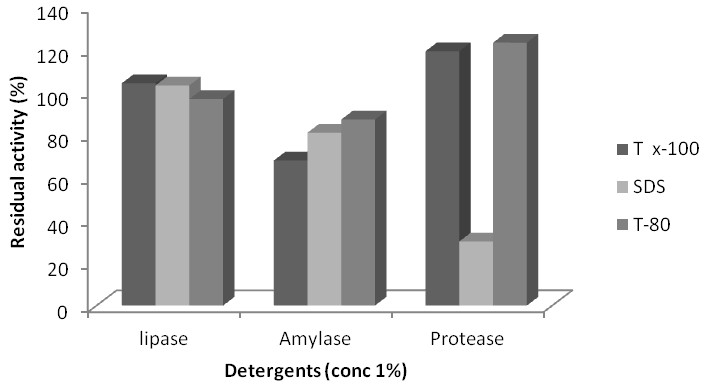
Figure 6 Effects of Detergents on thelipolytic, amylolytic and proteolytic activity of isolate Lysinibacillus sphaericus KC5-MRL.
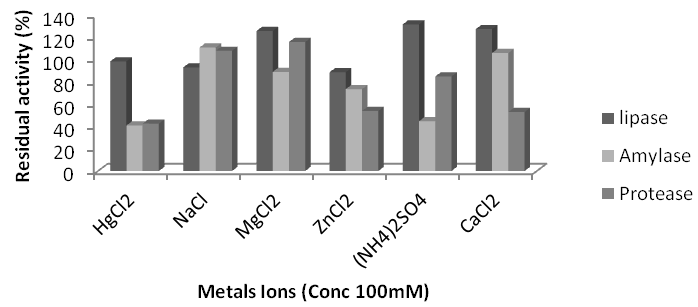
Figure 7 Effects of metals ions on lipase, Amylase and protease activity of isolate Lysinibacillus sphaericus KC5-MRL.
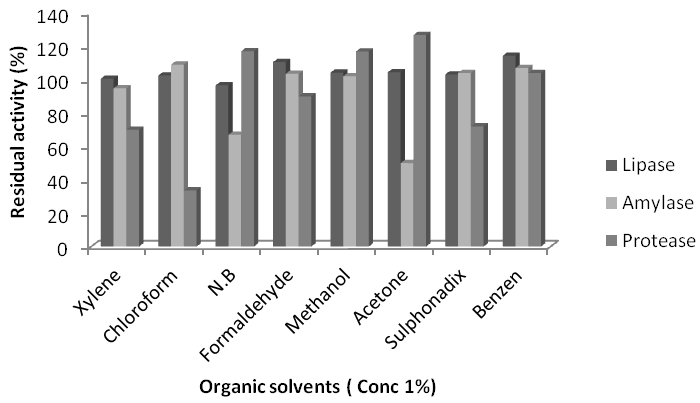
Figure 8 Effects of Organic solvents on Lipase, amylase and protease activity of isolate Lysinibacillus sphaericus KC5-MRL.
The study has been supported by the department of Microbiology Quaid-I-Azam University Islamabad, Pakistan.
The authors declare that there is no conflict of interest.

©2021 Afzal, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.