Journal of
eISSN: 2469 - 2786


Research Article Volume 4 Issue 2
1Department of Biochemistry, Ahmadu Bello University, Nigeria
2Department of Biological Sciences, Nigerian Defence Academy, Nigeria
3Forestry Research Institute of Nigeria (FRIN), Nigeria
Correspondence: Maryam Sani Lawal, Department of Biochemistry, Ahmadu Bello University, Zaria, Kaduna, Nigeria, Tel +2348038675358
Received: November 28, 2016 | Published: February 6, 2017
Citation: Lawal MS, Sani AM, Abdullahi UY, et al. Antimicrobial resistance pattern and plasmid profile of diarrheal salmonallae and shigallae bacteria isolated from clinical samples in Kaduna metropolis, Nigeria. J Bacteriol Mycol Open Access. 2017;4(2):41-45. DOI: 10.15406/jbmoa.2017.04.00084
Salmonallae and Shigellae bacteria of the family Enterobacteriaceae has been implicated as the causal agent of many gastro-intestinal disorders in man and animal. To determine the antimicrobial resistant pattern and plasmid profile of different resistant Salmonellae and Shigellae bacteria in Kaduna metropolis, Kaduna state, Nigeria, a total of fifty eight (58) isolates of clinical were collected randomly from diarrheal patients in the metropolis and tested against ten (10) commonly used antimicrobial agents using the Kirby-Bauer disc diffusion method. Plasmid DNA was extracted using FosmidMAX DNA purification kit and separated by agarose gel electrophoresis. Results indicated a varying degree of resistance to the different drugs tested, however, all Salmonella and Shigella isolates were found to be sensitive to at least one of the ten (10) drugs tested. Augmentin, Amoxicillin and streptomycin were found to be most resisted with no effect on any of the isolates while Chloramphenicol was the most effective with 25mm±1.00 and 23mm±1.00 against Salmonella and Shigella respectively. Plasmid profile revealed 31% success, suggesting no correlation between presence of plasmid and antibiotic resistance in the isolates. It was concluded that isolates might have harboured the presence of multi-drug resistance in their chromosomal genes rather than in plasmids.
Keywords: salmonellae, shigellae, antibiotic resistance pattern, plasmid profile, agarose gel electrophoresis, chloramphenicol, diarrhoea, impermeability, kirby-bauer disc diffusion method
Diarrhoea has been defined as the passage of loose, watery stool occurring more than three times in one day or the passage of loose or liquid stools more frequently than is normal for the individual.1 The disease may be mild and lasting a day or two and self-resolves without any special treatment. However, prolonged diarrhoea can be a sign of other problems. Diarrhea can cause dehydration, which is particularly dangerous in children and the elderly, and it must be treated promptly to avoid serious health problems. Enteric pathogens implicated in cases of diarrhoea may be bacterial, viral, fungal, parasitic or protozoal. Another infectious agent which has been implicated particularly in food-related infection is prion.2
Species of the genus Shigella and Salmonella are among the bacterial pathogens most frequently isolated from patients with diarrhoea. Five to fifteen percent of all diarrheal episodes worldwide can be attributed to an infection with Shigella, including 1.1 million fatal cases.3 Worldwide estimates of non typhoidal salmonellosis range from 200 million to 1.3 billion, with an estimated death toll of 3 million each year.4 The diarrheal diseases are usually characterized by blood, cramping, abdominal pain, fever, nausea, and vomiting.5
Resistance of pathogenic organisms to antibiotics is an increasing problem to the treatment of most microbial infection. The rapid dissemination of drug-resistant bacteria has been identified as a global problem that seriously complicates the treatment of human infections.6 However some of the factors reported to contribute to this increase, include high use of antimicrobial agents in humans and animals resulting in pressure for selection of resistant bacteria, the capacity of bacteria to disseminate antimicrobial resistance genes to other bacteria mainly by mobile genetic structures, and the facility of dissemination of resistant bacteria in different ecosystems.7 Antimicrobial resistance associated with specific antimicrobials occur in a number of ways. The various mechanisms of antimicrobials inactivation leading to the emergence of multiple drug resistance pathogens have been reported.8,9 Some of these include, reduced uptake, impermeability or efflux mechanisms for antimicrobial, drug degradation (enzyme attack) and or modification of specific target sites by the organism. Alternatively, organisms modify specific sites where antimicrobials normally act by duplication of the target site with a site that is non-susceptible thus causing a ‘‘bypass’’ of the antibiotic sensitive step.10,11 Genes coding for antibiotic resistance and virulence may share common features of being located in the bacterial chromosome, as well as on plasmids. In addition, gene clusters may form resistance or pathogenicity islands which are transferred by mobile elements (such as integrons or transposons) or phages.12
Integrons are a class of novel, naturally occurring mobile genetic elements that can capture antimicrobial resistance and other genes and promote their transcription.13,14 Integrons have been recognized as a mechanism for acquiring multiple antimicrobial resistances in some organisms. Recently, Class 1 type integrons are considered the most common in clinical isolates.15,16 Resistance genes can be integrated in the form of cassettes, a reaction which is catalysed by the integron encoded integrase (int1) gene at the att1 site.17,18 In many pathogenic bacteria, plasmids frequently carry antibiotics resistance encoding genes allowing bacteria to survive antibiotic treatment besides encoding virulence factors. This study was therefore aimed at investigating the antibiotic susceptibility pattern of diarrhea associated Sallmonella spp and Shigella spp, and to determine the possible role of plasmids in the resistance response of the isolates to commonly used antibiotics in Kaduna metropolis, Nigeria.
Collection and maintenance of test organisms
Pure cultures of bacteria(Salmonella spp, and Shigella spp,) isolated from clinical specimen were obtained from the Microbiology Department of Ahmadu Bello University Teaching Hospital Zaria, Yusuf Dan-tsoho Memorial Hospital, National Institute For Trypanosomiasis Research and Gold Bond Medical Laboratories, Kaduna. The organisms were maintained on Nutrient agar slants at 4˚C and were routinely sub-cultured during storage.
Identification of isolates
To confirm the identity of the clinical samples obtained from the health institutions they were subjected morphological and biochemical characterisation outlined in the Bergey’s manual of Determinative Bacteriology.19
Antibiotic susceptibility screening
The sensitivity/resistance of the bacterial strains to antibiotics was carried out using Kirby-Bauer disc diffusion method. A multi disc containing Augmentin (30microgram), Ciprofloxacin (10micrograms), Septrin (30micrgramms), Chloramphenicol (30microgramms), Sparfloxacin (10microgramms), Amoxicillin (30microgramms), Gentamycin (10microgramms), Pefloxacin (30microgramms), Tarivid (10micrgramms) and Streptomycin (30microgramms) was employed.
Isolation of plasmids
The isolation of plasmids from each of the bacteria culture was done using the FosmidMAX DNA Purification kit. Briefly, the pure colonies of drug-resistant bacteria isolated from the susceptibility studies earlier were grown in nutrient broth for 24h and thereafter harvested by centrifugation (1000 rpm). The supernatant was discarded and the residue containing the bacteria was harvested. The bacteria cells were re-suspended in 300μl of TENS solution and 3M sodium acetate to and spun for 2min. Supernatant containing plasmid was transferred to an empty sterile test tube, and spurned again for 2 min and the supernatant discarded. Residue (Pellets) obtained were rinsed twice in 1ml of 70% ethanol, dried under vacuum and re-suspended in 30μl of Tris EDTA buffer. The plasmid-containing preparation was then stored in a refrigerator at 4˚C until characterized.
Characterization of plasmids using agarose gel electrophoresis
Submerged horizontal agarose electrophoresis was employed to determine the size and fragments of the isolated plasmids according to a previously established method. Briefly, prepared plasmid mixture was electrophoresed using 1% agarose gel concentration in TBE running buffer (540g Tris, 225g Borate, 41.5g EDTA in water). A molecular ladder was used as a molecular weight marker, bromophenol was employed as the tracking dye. Electrophoresis was carried out at 60 mA and 220V for 2 hours. The gel was viewed on ultraviolet plate using protective goggles. Photomicrograph of the gel was generated to show the size, lane and profile of the plasmids. The molecular size of each plasmid was determined by comparison with the molecular ladder, and distances travelled.
Morphological and biochemical characteristics of isolates
The morphological and biochemical characteristics of the isolates obtained from clinical samples are presented in Table 1.
Phenotypic Traits and Tests |
Appearances and Responses of Bacterial Isolates |
|
Salmonella spp. |
Shigella spp. |
|
Morphology |
Smooth, Colourless, Colonies 2-4mm |
Smooth, Colourless, Colonies 2-3mm |
Shape |
Rod |
Rod |
Gram reaction |
-ve |
-ve |
Glucose |
+ve |
+ve |
Sucrose |
+ve |
+ve |
lactose |
-ve |
-ve |
Oxidase |
-ve |
-ve |
Indole |
-ve |
-ve |
Hydrogen Sulphide |
+ve |
-ve |
Gas production |
+ve |
+ve |
Ornithine |
+ve |
+ve |
Urea |
-ve |
-ve |
Citrate |
+ve |
-ve |
Motility |
motile |
non-motile |
Triple sugar iron(butt) |
A |
A |
Triple sugar iron (slant) |
K |
K |
Table 1 Morphological and biochemical characterisation of Salmonella spp and Shigella spp isolates from patients with diarrhoea
+ve: Positive; -ve: Negative; A: Acidic; K: Alkaline
Susceptibility testing of conventional antibiotics against Salmonella and Shigella species
It can be deduced from the result that while Augmentin, Amoxicillin and streptomycin were the most resisted antibiotics, having no effect on any of the isolates exposed to them (Table 2), Chloramphenicol was the most effective with 25±1.00 and 23±1.00 against Salmonella and Shigella respectively. Followed by Sparfloxacin (15.33±0.58) for Shigella and Salmonella with 21.00±1.00. Drug resistance was also observed for Gentamycin, perfloxacin and Tarivid. The result of the susceptibility of antibiotics presented a varying degree of resistance to the different drugs used against the different organisms but in all, from the result of antibiotics susceptibility tests, all isolates were sensitive to at least one of the ten antibiotics and resistant to more than three they were tested against. The result were labeled Resistant (R), Intermediate (I) and Susceptible (S) after being compared to the approved performance standards for antimicrobial susceptibility testing.20
Zones of Inhibitions (mm) |
||
Antibiotics |
Shigella |
Salmonella |
Chloramphenicol |
23.00 ± 1.00(S) |
25.00 ± 1.00(S) |
Sparfloxacin |
15.33 ± 0.58(I) |
21.00 ± 1.00(S) |
Septrin |
7.17 ± 0.29(R) |
14.67 ± 0.58(I) |
Ciprofloxacin |
20.50 ± 3.04(S) |
21.00 ± 1.00(S) |
Amoxicillin |
0.00 ± 0.00(R) |
0.00 ± 0.00(R) |
Augmentin |
0.00 ± 0.00(R) |
0.00 ± 0.00(R) |
Gentamycin |
0.00 ± 0.00(R) |
8.33 ± 0.58(R) |
Pefloxacin |
0.33 ± 0.58(R) |
0.00 ± 0.00(R) |
Tarivid |
4.83 ± 0.29(R) |
7.00 ± 1.00(R) |
Streptomycin |
0.00 ± 0.00(R) |
0.00 ± 0.00(R) |
Table 2 Zones of inhibition of antibiotics with Salmonella spp and Shigella spp
R: Resistant; I: Intermediate; S: Susceptible
Drug resistance of bacteria to antibiotics has been attributed to the misuse and overuse of antibiotics as well as the possession of drug resistance plasmids.21 Resistance is a natural biological response of microbes to antimicrobials and is currently a worrisome scenario affecting many parts of the world.22,23 Apart from intrinsic resistance, gene transfer and mutation are among the underlying mechanisms involved in the development of antimicrobial resistance by microbes.22 Several factors contribute to resistance by pathogens causing diarrhea in the setting of a developing country like Nigeria. These include frequent overuse, misuse and factors related to the potency and quality of antimicrobials and the distribution of resistant strains.22,24 In addition, syndromic diagnosis and diagnostic imprecision usually force physicians to opt for broad spectrum antibiotics such as amoxicillin and tetracycline, over prescribing; and less antibiotic diversity which lead to the emergence and spread of antimicrobial resistance.25 Fortunately, there seems to be limited resistance to the drugs chloramphenicol, ciprofloxacin and sparfloxacin.
Plasmid isolation
A total of 58 isolates were prepared for plasmid isolation, of which 30(51.7%) were Salmonella isolates and 28(48.3%) were Shigella isolates. Eight (26.67%) of the Salmonella isolates contained plasmids and Ten (35.7%) of the Shigella isolates also contained plasmids (Figures 1-4). The higher percentages (73.33% and 64.3%) of both Salmonella and Shigella organisms showed absence of plasmids (Figures 1 & 2), some of isolates of Salmonella and Shigella contained a heterogeneous population of plasmids ranging between the sizes of 200bp-2000bp. These were found resistant to three or more antibiotics. Antibiotic resistant Shigella spp and Salmonella spp have been reported to carry a heterogeneous population of plasmids, in an antimicrobial study on Salmonella spp and Shigella spp. Their isolates showed presence of varying number of plasmids even though the results of that study showed no clear or distinct correlation between plasmids and antibiotic resistance26 as in this study. In a similar study, multi-drug resistant strains of Shigella which were found resistant against three or more antibiotics contained a number of plasmids varied from one to seven27 and could reach up to ten.28

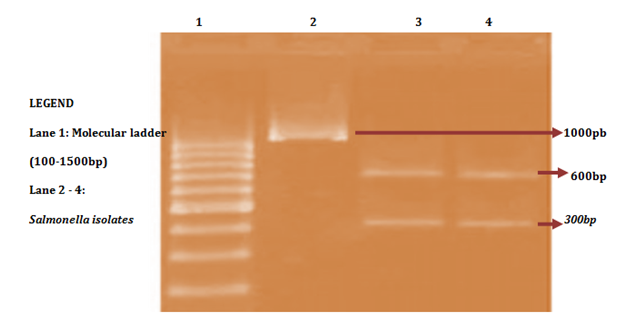

Antibiotic resistant bacteria are able to transfer copies of DNA that code for a mechanism of resistance to other bacteria even distantly related to them, which then are also able to pass on the resistance genes and so generations of antibiotics resistant bacteria are produced.29 This process is called horizontal gene transfer. Plasmid profile analysis is a useful tool in epidemiological studies dealing with enteric infections.
These results indicate a high probability of identity between the R-plasmids found in Shigella which usually encoded resistance to, amoxicillin, tetracycline, chloramphenicol, sulfamethoxazole- trimethoprim and co-trimoxazole. The cause of the increase in R factor- carrying bacteria is due to the selective pressure caused by antibiotics. It has been shown that the use of antibiotics in animals greatly increase the pool of R factor- carrying bacteria in the environment. It seems likely that the use of antibiotics for other non-medical purposes also helps the increase of the reservoir of R factors.30 There are large numbers of antimicrobial agents such as penicillin, cephalosporin, tetracycline, spectinomycin, chloramphenicol, fusidic acid, sulfonamides, heavy metal and others for which plasmid-mediated antibiotic resistance has been reported.
In conclusion, except for Chloramphenicol, ciprofloxacin and sparfloxacin for which both Salmonella and Shigella isolates were susceptible, a high level of antimicrobial resistance was detected. Notably, the organisms seem to have developed complete resistance to Augmentin, streptomycin, pefloxacin and amoxicillin. We assert that ciprofloxacin and chloramphenicol may be drugs of choice for treating diarrheal attacks by these organisms in the study area. In the study, the result obtained indicates that there appears to be no correlation between plasmid and antibiotic resistance of Salmonella and Shigella though there might be exceptions.
None.
The author declares no conflict of interest.

©2017 Lawal, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.