Journal of
eISSN: 2469 - 2786


Research Article Volume 3 Issue 2
1Entomology Research Institute, India
2International Scientific Partnership Program, King Saud University, Saudi Arabia
3Department of Plant Biology and Biotechnology, Loyola College, India
4Department of Botany and Microbiology, King Saud University, Saudi Arabia
Correspondence: Ignacimuthu S, Entomology Research Institute, Loyola College, Chennai - 600 034, Tamil Nadu, India, Tel +91 44 2817 8348, Fax +91 442817 5566
Received: October 19, 2016 | Published: December 7, 2016
Citation: Devadass BJ, Paulraj MG, Ignacimuthu S, et al. Antimicrobial activity of soil actinomycetes isolated from Western Ghats in Tamil Nadu, India. J Bacteriol Mycol Open Access. 2016;3(2):224-232. DOI: 10.15406/jbmoa.2016.03.00059
Some actinomycetes were isolated from the soil samples collected from Kanyakumari, Tirunelveli, Dindigul, Coimbatore and the Nilgiri districts of Western Ghats in Tamil Nadu, India. They were screened for antibacterial and antifungal activities. Among the 150 strains of actinomycetes isolated from the soil samples, 21 isolates were active against more than 4 tested pathogens. Five isolates were potentially active against more than 8 tested pathogens. These five isolates were identified by 16S rDNA sequence anaylsis and the sequences were submitted to GenBank. These five isolates were sensitive to vancomycin. They all grew well at 20°C. Two isolates namely ERI-15 and ERI-17 showed very high inhibitory activity against E. faecalis and C. albicans. These can be probed further for their use in controlling pathogens.
Keywords: actinomycetes, antimicrobial activity, broth micro dilution method, mic50, 16s rdna sequencing
Actinomycetes are an extensive and diverse group of Gram-positive, aerobic and mycelial bacteria. They are one of the major communities of the microbial population present in the soil and they play an important ecological role in soil cycles. Many actinomycetes are economically important since they are the producers of antibiotics, vitamins and enzymes.1-6 They have been mostly isolated from marine habitats as a consequence of their adaptation to the saline environment. The most commonly isolated actinomycete genera are Streptomyces and Micromonospora and the majority of metabolites isolated are derived from a relatively limited pool of organisms.7 The number of novel secondary metabolites continues to increase in pharmacological and biological screenings, indicating that new producer strains and species of Streptomyces with great potential in biotechnology exist in nature.8-10
The development of resistance to multiple drugs is a major problem in the treatment of infections by pathogenic microorganisms. The increasing trend in the development of multi-drug resistance in microbes is mainly due the increased antibiotic use by humans and animals, global trade and human movement.11 This is presently an urgent focus of research and new bioactive compounds are necessary to combat these pathogens. This fact has made Streptomyces spp. the most used bacteria for the manufacture of active pharmaceutical compounds. It is essential that the search for new antibiotics need to be continued because of the many toxic effects of currently used compounds and the emergence of resistant pathogens.
The aim of the present study was to isolate members of actinomycetes from forest soil from Western Ghats and to investigate their antagonistic properties against bacteria and fungi.
Sample collection
Soil samples were collected from five districts namely Kanyakumari, Tirunelveli, Dindigul, Coimbatore and the Nilgiris of Western Ghats in Tamil Nadu. Approximately 500 gm of the soil samples were collected in sterile bags using a spatula. Totally 67 samples were collected from these regions and were further processed by following the method of Hayakawa et al.12 The collected soils were passed through a 2 mm mesh sieve, air-dried at room temperature for 7 days and used for actinomycetes isolation.
Isolation and selection of bioactive actinomycetes
Ten milliliters of sterile distilled water were added to 1.0 gm of dried soil, vortexed vigorously for 1 min and allowed to stand for 5 min. Serial 10-fold dilutions from 10-1 to 10-5 of the soil suspensions were prepared in sterile distilled water and 0.1 ml volumes from 10-3 to 10-5 were plated on Starch Casein Agar plates (Hi-media), containing cycloheximide (40μg/ml) (Hi-media) and Nalidixic acid (100μg/ml) (Hi-media). The plates were incubated at 28˚C for 7 to 16 days or until colonies were formed. The selection of the actinomycetes colonies were carefully observed for the typical morphology as per Bergey’s manual and as per the proceedings of the international Streptomyces project published in the international Journal of systemic bacteriology by.13
The test microorganisms used in the present study were procured from American type culture collection (ATCC) and Microbial type culture collection (MTCC) Chandigarh, India. Nine bacteria namely Bacillus subtilis (MTCC441), Staphylococcus aureus (ATCC 25923), Staphylococcus epidermidis (MTCC 3615), Enterococcus faecalis (ATCC 29212), Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), Klebsiellapneumoniae (ATCC15380), Xanthomonascampestris PV oryzae and Erwiniaamylovora MTCC 2760 and seven fungi viz., Aspergillusniger (MTCC1344), Aspergillusflavus, Botrytis cinerea, Curvularialunata 46/01, Scopulariopsissp. 101/01, Trychophytonsimii 110/01 and Candida albicans (MTCC 227) were chosen for screening experiments.
Preliminary screening of actinomycetes for antibacterial and antifungal activities
The preliminary antibacterial screening was performed for all the isolated cultures by cross streak method, against the selected Gram positive and Gram negative bacteria and fungi, which are medically and agriculturally important pathogens. The isolated actinomycetes were inoculated as a line of streak at the centre of the Modified Nutrient Glucose Agar (MNGA) plates as described by Balagurunathan.14 The test cultures were inoculated by cross-streak method i.e. line of streak perpendicular to the actinomycetes growth. The plates were incubated at 37˚C for 24 h in the incubator for bacteria and at 28˚C for 24-48 h for Candida albicans. The inhibitory action of the actinomycetes was verified by the absence of growth around the antibiotic producing actinomycetes and the results were recorded.
Media optimization
Five isolates namely ERI-11, ERI-14, ERI-15, ERI-17 and ERI-28 were identified as the most active isolates. Growth characteristics of these five isolates were studied on different international Streptomyces project media (ISP-2 to ISP-5). The best medium was used for growing the isolates for crude extract preparation from their culture broth.
Production of crude extract
The potential cultures with good antibiotic activity were further chosen for crude extract preparation for secondary screening. About 1000 ml of optimized ISP-2 was distributed in approximately 200 ml in five Erlen Meyer flasks (1000 ml volume) and 5µl of spore suspension of each active culture was inoculated into respective flasks under sterile aceptic conditions in Laminar air-flow. The inoculated flasks were incubated at room temperature for 7 days. On the 7th day the fermented broth was centrifuged in 50 ml centrifuge tube at 10000 rpm for 5 min. The supernatant was collected and filtered in order to avoid any cell debris. The fermentation broth was subjected to sequential extraction using hexane, chloroform and ethyl acetate in a 1000 ml separating funnel. The organic phase of the supernatant was dried under reduced pressure using rotary vacuum evaporator and concentrated extracts were collected, dried and used for bioassay studies.
Secondary screening of active isolates by disc diffusion method
One milligram of the crude extract was dissolved in 100µl sterile DMSO and 400µl of sterile double distilled de-ionized water was added and mixed thoroughly. From this, 30µl of the treatment solution was impregnated onto sterile discs (6 mm; Himedia, Mumbai) under aseptic conditions. The discs were allowed for complete drying in the oven at 45˚C. Sterile Muller Hinton agar plates were prepared and the pathogenic bacteria (five Gram positive and four Gram negative) were swabbed on to the surface of the agar plates under aseptic conditions. The discs impregnated with the extracts were placed on the plates and incubated at 37˚C for 24 h. The zone of inhibition was recorded for all the extracts against all pathogenic bacteria. DMSO was used as solvent control and streptomycin was used as reference control.
For antifungal bioassay experiments, the extracts were screened against seven fungal organisms. The spore suspensions of the selected fungal pathogens were swabbed on to Muller Hinton Agar plates under aseptic conditions. The discs impregnated with the extract solutions were placed on to the agar plates and incubated at room temperature for 48-72h. The zone of inhibition was recorded for all the extracts against all test fungi. DMSO was used as solvent control and ketoconazole was used as reference control.
Secondary screening by well diffusion method
Sterile Muller Hinton Agar plates were prepared and the pathogenic bacteria and fungi were swabbed on to the surface of the agar plates under aseptic conditions. The well was prepared in agar plates using a sterile cork-borer (6 mm diameter). A volume of 100µl of the crude extract was dispensed into the well carefully and allowed for 2 h for diffusion. After 2 h of inoculation, the plates were incubated for 24h at 37˚C and the zone of inhibition around the well was measured and recorded. DMSO was used as solvent control. Streptomycin (for bacteria) and Ketoconazole (for fungi) were used as reference control.
Determination of the MIC was performed according to the standard reference method.15 MIC was calculated against four bacteria namely E. coli, P. aeruginosa, S. aureus and E. faecalis and one fungus namely C. albicans. Crude extract was dissolved in 10% DMSO. The initial concentration of the crude extract was 1mg ml-1 and this initial test concentration was serially diluted. Each well was inoculated with 107 CFU ml-1 of bacteria suspension (5ml) and 104 spore ml-1 of fungi (5ml), respectively. Fluconazole and ciprofloxacin were used as positive controls for fungi and bacteria, respectively. The plates were incubated at 27˚C up to 5 days for fungi and 24 h at 37˚C for bacteria. The MIC of extract was found as the lowest concentration of the compound inhibiting the visual growth of the organisms.
Physiological and biochemical characteristics of active isolates
The growth of the five most active isolates at different temperatures, pH and salinity, utilization of carbon sources such as L-Arabinose, Cellulose, Fructose, D-Glucose, Myo-Inositol, D-Raffinose, L-Ramnose, Sucrose, D-Mannitol and D-Xylose and Degradation of Aesculin, Hypoxanthine, Tween 80, Xanthine, Casein, Starch, Gelatin, Chitin, Urea and L-Tyrosine were studied.
Micro morphology of active isolates by Scanning Electron microscopy (SEM)
The ISP-4 medium-grown 14-days old cultures of each actinomycete were observed by a simple spore print technique in which Formvar-covered copper grids (200 mesh) (Shawinigan Resin Corporation, Springfield, Massachusetts) were gently pressed to the sporulating surface of the organisms and the resulting samples were dried at the critical point of liquid CO2. The samples were sputter-coated with gold (1-3 run) and examined under a scanning electron microscope (Hitachi, SEM, S-3400N, Type II, Tokyo, Japan).
Antibiotic resistance/susceptibility tests
The antibiotic sensitivity of five active isolates was tested against 12 antibiotics namely Cephotoxime, Chloramphenicol, Ciprofloxacin, Clindamycin, Co-trimoxazole, Erythromycin, Gentamycin, Linzolid, Oxacillin, Penicillin G, Tetracycline and Vancomycin.
Molecular identification by 16s rDNA sequence analysis and phylogenetic tree
The molecular identification of five active isolates was done by 16s rDNA sequence analysis. The total DNA was extracted by the method of.16 The 16S rDNA was amplified using universal primers 27f (50-AGAGTTTGATCMTGCCTCAG-30) and 1492r (50-TACGGYTACCTTGTTACGACTT-30). The 16S rDNA gene sequences of five isolates were submitted to GenBank and were provided with Nucleotide sequences accession numbers for the same. Molecular Phylogenetic analysis was done by Maximum Likelihood method. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model.17 Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 675 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 software.18
Sample collection and isolation of actinomycetes
Totally 67 soil samples were collected from 18 different locations within five districts along the Western Ghats of Tamil Nadu. Twenty nine soil samples were collected from Kanyakumari district; 5 samples were from Tirunelveli district; 11 samples were from Dindigul district; 11 samples were from Nilgiris district and 11 samples were from Coimbatore district. A total of 733 actinomycetes were selected from Starch-Casein Agar plates and screened for antimicrobial properties. Most of these isolates showed typical morphology of Streptomyces, with branched and non-fragmented substrate mycelia, abundant aerial hyphae and short or long spore chains with or without pigmentation. About 434 actinomycetes were isolated from Kanyakumari district; 62 actinomycetes were from Tirunelveli district; 43 actinomycetes were from Dindigul district; 66 actinomycetes were from Nilgiris district and 128 actinomycetes were from Coimbatore district. Out of the 733 strains isolated, 700 strains were further taken for pure culturing and maintenance since some of the strains did not survive due to autolysis during pure culturing.
The well grown cultures were picked up for slant transfer. Various morphological features such as colony morphology, color of the aerial and substrate mycelium, spore morphology and soluble pigments production were observed. Streptomycetes colonies were carefully selected for slant transfer after screening by their appearancesas chalky, leathery, lichenoid, discrete, or butyrous.
Preliminary screening bioassays
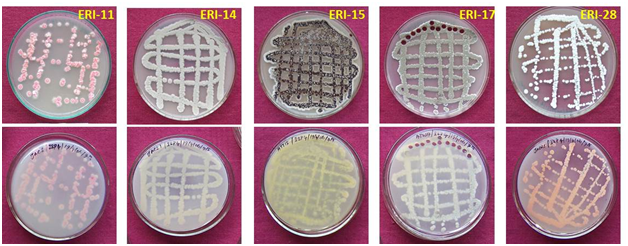
The cross streak method was used extensively against wide range of test pathogens to determine the potential antimicrobial actinomycetes (Figure 1). Preliminary screening experiment clearly showed that out of the 700 actinomycetes, only 150 actinomycetes isolates possessed antimicrobial property. About 21 isolates were active against most of the tested microorganisms. Five isolates showed very high antimicrobial activity against more than eight pathogenic microbes. These five active isolates were: three isolates from Kanyakumari district viz., ERI-11, ERI-14 and ERI-15, one isolate (ERI-17) from Tirunelveli district and one isolate (ERI-28) from Nilgiri district. The morphology of the five isolates is presented in Figures 2 & 3. Majority of tested actinomycetes was active against S. aereus, E.coliand C. albicans.
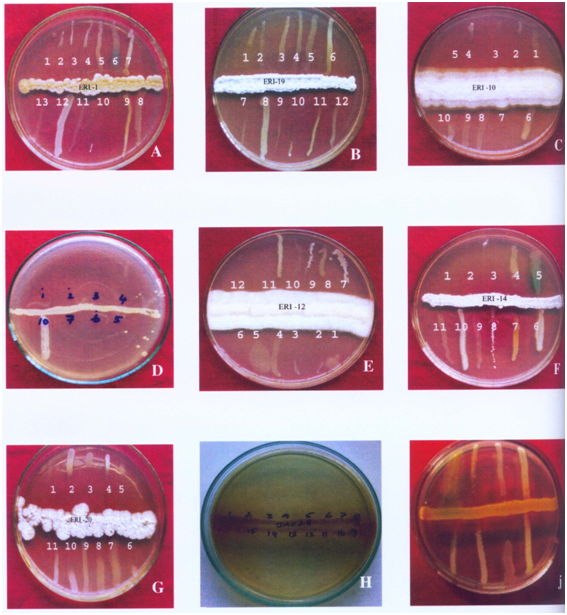

Secondary screening by well diffusion method: Table 1 shows the agar well diffusion assay results. The results showed that ERI-15 and ERI-17 were active against 15 microbes. ERI-15 inhibited growth of 10 pathogenic microbes above 10 mm level. ERI-28 was active against 14 pathogenic microbes and it showed zone of inhibition of more than 10 mm against seven pathogens. ERI-11 and ERI-14 were active against 11 and 12 pathogens respectively.
Sl. No |
Test Organisms |
Zone of Inhibition (mm) |
|||||
ERI-11 |
ERI-14 |
ERI-15 |
ERI-17 |
ERI-28 |
*Positive control |
||
1 |
Bacillus subtilis MTCC 441 |
6 |
6 |
11 |
8 |
9 |
13 |
2 |
S. aureus ATCC 25923 |
6 |
- |
10 |
6 |
7 |
12 |
3 |
S. epidermidis MTCC 3615 |
9 |
8 |
12 |
10 |
12 |
15 |
4 |
Ent. faecalis ATCC29212 |
8 |
6 |
13 |
11 |
12 |
13 |
5 |
E. coli ATCC 25922 |
- |
- |
6 |
7 |
- |
16 |
6 |
P. aeruginosa ATCC 27853 |
- |
- |
- |
- |
- |
15 |
7 |
K. pneumoniae ATCC 15380 |
- |
8 |
9 |
7 |
9 |
14 |
8 |
Xanthomonas sp. |
7 |
7 |
9 |
8 |
10 |
13 |
9 |
E. amylovora MTCC 2760 |
9 |
9 |
9 |
8 |
9 |
14 |
10 |
C. albicans MTCC 227 |
9 |
10 |
12 |
10 |
9 |
12 |
11 |
A. niger MTCC 1344 |
8 |
8 |
13 |
11 |
12 |
16 |
12 |
A. flavus |
8 |
9 |
12 |
10 |
12 |
15 |
13 |
Botrytis cinerea |
7 |
8 |
14 |
12 |
11 |
12 |
14 |
Curvularia sp. 46/01 |
8 |
8 |
13 |
8 |
10 |
14 |
15 |
Scopulariopsis sp. 101/01 |
- |
7 |
10 |
6 |
7 |
16 |
16 |
T. simii110/01 |
- |
- |
6 |
7 |
6 |
15 |
Table 1 Antimicrobial activity of ethyl acetate extract of fermented broth obtained from five potential actinomycetes against pathogenic bacteria and fungi by well diffusion method
- no activity; *Streptomycin for bacteria and Ketoconazole for fungi
Secondary screening by disc-diffusion method: Hexane, chloroform and ethyl acetate extracts of culture filtrates of five active isolates were screened for antimicrobial activity. Hexane extract had no antimicrobial activity against the tested microbes and chloroform extract showed very mild activity. But ethyl acetate extract presented very high antimicrobial activity against all the nine bacterial and five fungal pathogens. The results are presented in Table 2. ERI-15 and ERI-28 were found to be very effective against more number of pathogenic microbes. Zone of growth inhibition above 20 mm was achieved against 10 and 9 microbes by ERI-15 and ERI-28 respectively. ERI-17 showed zone of inhibition above 20 mm against 6 pathogens. The antimicrobial activity of ethyl acetate extracts of ERI-15 and ERI-28 was almost comparable with standard drugs (positive controls).
Sl. No |
Test Organisms |
Zone of Inhibition (mm) |
|||||
ERI-11 |
ERI-14 |
ERI-15 |
ERI-17 |
ERI-28 |
*Positive control |
||
1 |
Bacillus subtilis MTCC 441 |
19 |
18 |
24 |
21 |
22 |
18 |
2 |
S. aureus ATCC 25923 |
18 |
15 |
23 |
19 |
20 |
17 |
3 |
S. epidermidis MTCC 3615 |
23 |
21 |
28 |
25 |
27 |
25 |
4 |
Ent. faecalis ATCC29212 |
22 |
20 |
28 |
24 |
27 |
23 |
5 |
E. coli ATCC 25922 |
9 |
7 |
13 |
9 |
11 |
20 |
6 |
P. aeruginosa ATCC 27853 |
8 |
7 |
11 |
8 |
9 |
19 |
7 |
K. pneumoniae ATCC 15380 |
8 |
6 |
12 |
10 |
11 |
18 |
8 |
Xanthomonas sp. |
15 |
6 |
18 |
16 |
22 |
20 |
9 |
E. amylovora MTCC 2760 |
16 |
7 |
19 |
17 |
19 |
19 |
10 |
C. albicans MTCC 227 |
18 |
12 |
26 |
21 |
18 |
20 |
11 |
A. niger MTCC 1344 |
15 |
14 |
27 |
25 |
28 |
22 |
12 |
A. flavus |
15 |
14 |
28 |
13 |
28 |
18 |
13 |
Botrytis cinerea |
14 |
16 |
30 |
26 |
25 |
23 |
14 |
Curvularia sp. 46/01 |
15 |
21 |
28 |
16 |
23 |
25 |
15 |
Scopulariopsis sp. 101/01 |
12 |
10 |
22 |
13 |
14 |
17 |
16 |
T. simii 110/01 |
11 |
9 |
14 |
12 |
16 |
18 |
Table 2 Antimicrobial activity of ethyl acetate extract of fermented broth obtained from five potential actinomycetes by disc diffusion method
- no activity; *Streptomycin for bacteria and Ketoconazole for fungi
Minimum inhibitory concentration
The active strains were tested against S. aureus, C. albicans, E.coli, P. aeruginosaand E. faecalis. The results clearly showed that both ERI-15 and ERI-17 were the most effective antimicrobials against the tested pathogens. ERI-15 recorded the lowestIC50 values of 0.33 and 0.37mg/ml against E. faecalis and C. albicans, respectively. The same strain also recorded the lowest IC99value of 0.72mg/ml against both E. faecalis and C. albicans (Table 3).
Pathogens |
MIC50 (mg/ml) |
MIC99 (mg/ml) |
||||||||
ERI -11 |
ERI -14 |
ERI- 15 |
ERI -17 |
ERI -28 |
ERI -11 |
ERI- 14 |
ERI -15 |
ERI -17 |
ERI- 28 |
|
E. coli |
2.22 |
2.5 |
1.13 |
2.2 |
>4.5 |
>4.5 |
3.7 |
2.23 |
3.87 |
>4.5 |
P. aeruginosa |
4.51 |
>4.2 |
2.12 |
4.2 |
2.23 |
>4.81 |
>4.5 |
3.91 |
4.8 |
3.24 |
S. aureus |
1.12 |
>4.2 |
0.69 |
1.32 |
2.52 |
1.93 |
>4.82 |
1.39 |
2.42 |
3.79 |
E. faecalis |
0.85 |
1.12 |
0.33 |
0.56 |
1.49 |
1.63 |
1.79 |
0.72 |
1.03 |
2.42 |
C. albicans |
0.71 |
0.86 |
0.37 |
0.63 |
2.71 |
0.98 |
1.24 |
0.72 |
1.31 |
4.31 |
Table 3 Minimum inhibitory concentrations of active isolates against pathogenic bacteria and fungi
Physiological and biochemical characteristics of active isolates\
Table 4 shows the growth and physico-chemical characteristics of the active five isolates. ERI-14, ERI-17 and ERI-28 grew in salinity up to 7%. All isolates showed growth at 20˚C. But at higher temperature (45oC), ERI-14 did not grow and other four isolates showed growth. In phenol and sodium azide conditions, none of the isolates grew. ERI-11 utilized all carbon sources tested except cellulose. ERI-17 could not utilize L-Arabinose, Fructose, Myo-Inositol, D-Mannitol and D-Xylose. D-Glucose, D-Raffinose and Sucrose were utilized by all five active isolates. Cellulose was utilized as carbon source by ERI-14, ERI-15 and ERI-17. Gelatin was found to be degraded by all the five isolates. Casein, starch and L-Tyrosine were degraded by four isolates namely ERI-14, ERI-15, ERI-17 and ERI-28. Chitin was degraded by ERI-14, ERI-15 and ERI-17.
Characteristics |
Active Isolates |
||||
ERI-11 |
ERI-14 |
ERI-15 |
ERI-17 |
ERI-28 |
|
Carbon source utilization |
|||||
L-Arabinose |
+ |
+ |
- |
- |
+ |
Cellulose |
- |
+ |
+ |
+ |
- |
Fructose |
+ |
+ |
+ |
- |
+ |
D-Glucose |
+ |
+ |
+ |
+ |
+ |
Myo-Inositol |
+ |
- |
+ |
- |
+ |
D-Raffinose |
+ |
+ |
+ |
+ |
+ |
L-Ramnose |
+ |
+ |
- |
+ |
- |
Sucrose |
+ |
+ |
+ |
+ |
+ |
D-Mannitol |
+ |
- |
+ |
- |
+ |
D-Xylose |
+ |
+ |
+ |
- |
+ |
Degradation |
|||||
Aesculin |
+ |
- |
+ |
+ |
+ |
Hypoxanthine |
+ |
- |
- |
+ |
+ |
Tween 80 |
+ |
+ |
- |
+ |
+ |
Xanthine |
+ |
- |
- |
- |
+ |
Casein |
- |
+ |
+ |
+ |
+ |
Starch |
- |
+ |
+ |
+ |
+ |
Gelatin |
+ |
+ |
+ |
+ |
+ |
Chitin |
- |
+ |
+ |
+ |
- |
Urea |
- |
- |
- |
- |
|
L-Tyrosine |
- |
+ |
+ |
+ |
+ |
Growth at/in |
|||||
20˚C |
+ |
+ |
+ |
+ |
+ |
45˚C |
+ |
- |
+ |
+ |
+ |
pH 10.0 |
+ |
- |
+ |
- |
- |
NaCl (7%) |
- |
+ |
- |
+ |
+ |
Phenol (0.1%, w/v) |
- |
- |
- |
- |
- |
Sodium azide (0.01%, w/v) |
- |
- |
- |
- |
- |
Table 4 Physiological and biochemical characteristics of five active isolates
+ growth; - no growth
Antibiotic resistant/sensitivity of active isolates
The antibiotic resistance/sensitivity test results obtained by disc diffusion method for all the five active isolates are presented in Table 5. The results showed that all the five isolates were resistant to penicillin, oxacillin, cephotaxime and co-trimoxazole. All were sensitive to chloroamphenicol, erythromycin and vancomycin.
Antibiotics (µg/disc) |
Active Isolates |
||||
ERI-11 |
ERI-14 |
ERI-15 |
ERI-17 |
ERI-28 |
|
Cephotoxime (30 µg) |
+ |
+ |
+ |
+ |
+ |
Chloramphenicol (30 µg) |
- |
- |
- |
- |
- |
Ciprofloxacin (5 µg) |
- |
+ |
+ |
- |
+ |
Clindamycin (2 µg) |
- |
- |
- |
- |
- |
Co-trimoxazole (25 µg) |
+ |
+ |
+ |
+ |
+ |
Erythromycin (15 µg) |
- |
- |
- |
- |
- |
Gentamycin (10 µg) |
- |
- |
- |
+ |
- |
Linzolid (30 µg) |
- |
+ |
- |
- |
- |
Oxacillin (1 µg) |
+ |
+ |
+ |
+ |
+ |
Penicillin G (10 U) |
+ |
+ |
+ |
+ |
+ |
Tetracyclin (30 µg) |
- |
- |
+ |
- |
+ |
Vancomycin (30 µg) |
- |
- |
- |
- |
- |
Table 5 Antibiotic sensitivity of five active isolates against selected antibiotics
+ resistant; - sensitive
Morphology and molecular characterization of Potential isolates
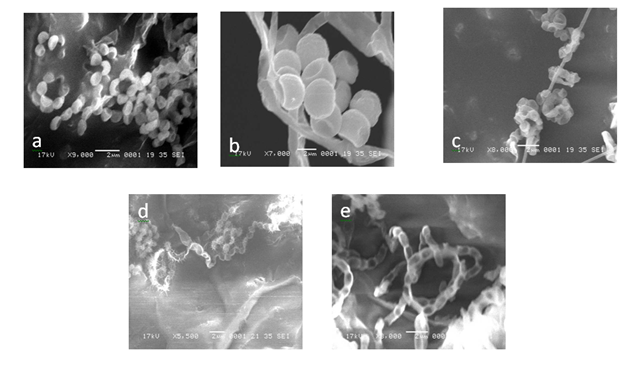
The scanning electron micrograph of active isolates showed the nature and structural differences of spore and hyphal morphologies between the five species. The typical characteristics of Streptomyces were seen and chains of spores were straight or coil (Figure 4). The size of the spores of the active actinomycetes ranged from 0.85 to 2.0µm and the spores were smooth for ERI-11, ERI-14 and ERI-28. The ERI-15 strain had warty spore coiled in chains of 5-10 spores. The spiny spores in chains of more than 10 numbers were found in ERI-17 and the retinaculum apertum of ERI-11 showed warty surface; ERI-14 and ERI-28 were smooth surface in nature but spores appeared as hanging bunch whereas ERI-28 showed chain like smooth spores. These observations were further confirmed in 16s rDNA sequence analysis. The results obtained from this sequence analysis confirmed ERI-11 as Actinomadura sp. which had 99% sequence similarity to Actinomadura sp. (EU274357); the nucleotide substitution was 0.005. The isolate ERI-14 was identified as Salinisporaarenicola, which had 99% similarity to the sequence of Salinispora sp. (GQ163717); the nuleotide substitution was 0.0015; this substitution was more relevant since this isolate was confirmed upto species level as Salinosporaarenicola. When compared with other S.arenicola the similarity was 96%. ERI-15 and ERI-17 were identified as Streptomyces sp. and had 99% similarity with Streptomyces sp.; ERI-17was 100% similar to Streptomyces rochei (FJ486442). The GenBank accession numbers for ERI-11, ERI-14, ERI-15, ERI-17 and ERI-28 were GU998794, JF836083, FJ705810, HQ127628 and JF836082, respectively.
The phylogenetic tree constructed with the five sequences of active isolates is shown in Figure 5. The tree with the highest log likelihood (-4551.5890) is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. ERI-15 and ERI-17 occupied the same clade; both were belonging to Streptomyces and were closely related. ERI-14 (Salinisporaarenicola) and ERI-28 (Micromonosporaechinospora) occupied the same clade.
Development of multidrug resistance in bacteria and fungi is really an obstacle in the treatment of pathogenic diseases. This creates an urgent need for new potent antibiotics to control the resistant pathogens.19 Actinomycetes, particularly in the genus Streptomyces, possess the ability to produce different biologically active secondary metabolites.20 The search for useful secondary metabolites from actinomycetes requires intensive screening of a large number of isolates obtained from unexplored soil samples. For this reason, soils were specifically collected from the forest regions. In the present study soil samples were collected from five different districts of Western Ghats in Tamil Nadu, India. This was based on the hypothesis that actinomycetes diversity might be influenced by the diversity of plant species as these bacteria will grow profusely in the humus and leaf litter layer.21
Since the Western Ghats are one of the important hotspot regions in the world due to a rich diversity of flora and fauna, our main focus was to isolate the actinomycetes from this region. Furthermore, different plants produce different types of secondary metabolites and some of these chemical compounds are toxic to soil microorganisms including actinomycetes. However, actinomycetes have developed adaptations to withstand such an adverse environment and produce their own secondary metabolites. In the present study 50 to 60 colonies were found per plate. Colonies selected from each plate were 5 to 20 based on colony appearance. Colonies having characteristic features such as powdery appearance with convex, concave or flat surface and color ranging from white, gray to pinkish and yellowish were selected. Colonies observed at 1st and 2nd day were eliminated because actinomycetes are considered as slow growers.22
In the present study, 20.5% of the total isolates showed antimicrobial property; but this differed from district to district as evident from the results obtained. The results of the screening revealed that more isolates were active against Gram positive bacteria than Gram negative bacteria. This might be due to the morphological differences between these two types of microorganisms. Gram negative bacteria have an outer lipopolysaccharide membrane; hence their cell wall is impermeable to lipophilic solutes, while porins constitute a selective barrier to the hydrophilic solutes.23 The Gram positive bacteria have only peptidoglycan layer, not an effective permeability barrier for the antibacterial agents. In the present study, 150 actinomycetes strains were isolated from 67 soil samples. The antimicrobial screening of different isolates against bacteria and fungi clearly showed that 21 isolates were active against more than 4 tested pathogens. From these 21 isolates, ERI-11, ERI-14, ERI-15, ERI-17 and ERI-28 were identified as the most effective isolates. The growth of these five effective isolates was not affected at a lower temperature of 20˚C and at a salinity level of less than 7% NaCl; at 45˚C, the growth of ERI-14 alone was affected and other four isolates grew at this higher temperature. At 7% salinity, ERI-14, ERI-17 and ERI-28 showed growth. Moncheva et al.24 have reported that six actinomycete strains isolated from Antarctic soil grew well at 25˚C and in 1.5% salinity.
Forest soils have rich biodiversity of actinomycetes. Previously Arasu et al.25 had isolated different actinomycetes strains from Western Ghats rock soil and reported a potential antimicrobial Streptomyces sp. which showed very low MIC of 0.25 mg/ml against Staphylococcus aureus, Staphylococcus epidermidis, Xanthomonas spp. and Candida albicans. Recently Ganesan et al.26 had isolated 106 soil actinomycetes from Western Ghats in Tamil Nadu and screened against pathogenic microbes. They reported that one isolate, FMS-20, was highly active against all tested pathogens. In another study, 356 Streptomyces isolates were obtained from soils in the Aegean and East Black Sea regions of Turkey and 36% of the isolates were found to be active against tested microorganisms.27 In our study, five actinomycetes were found to be the most effective among which three isolates namely ERI-15 (Streptomyces sp.), ERI-17 (Streptomyces rochei) and ERI-28 (Micromonosporaechinospora) were more potent. Streptomyces is the largest genus of action bacteria and nearly 615 species of Streptomyces have been described.28 Also it is the largest antibiotic-producing genus in the microbial world.29 Several Streptomyces spp. are generally acting as protective mutualistic symbionts in human beings and in other organisms. These symbionts provide antibiotics to protect the hosts from pathogens.30 However the symbiotic Streptomyces spp. can also cause opportunistic infections like actinomycetomas in humans.31,32 So it is very essential to study their resistance or susceptibility status against an array of commercial antibiotics. In the present study12 antibiotics were screened against the five active isolates. All five active isolates were susceptible to chloramphenicol, clindamycin, erythromycin and vancomycin. Resistance was recorded against cephotoxime, co-trimoxazole, oxacillin and penicillin. In a previous study, it was reported that 87 actinomyces species were susceptible to penicillin and amoxicillin.33 Wright34 proposed that soil offers a reservoir of antibiotic resistance genes known as‘soil resistome’ which cause the evolution of drug resistance in pathogenic bacteria.
In general the crude extracts of actinomycetes are not toxic or have very least toxicity to mammals.35 So the active isolates of the present study can be probed further for their use in the treatment of pathogenic organisms.
Totally 67 soil samples were collected from Western Ghats in Tamil Nadu and 733 strains of actinomycetes were isolated. Around 150 (20.4%) isolates showed antimicrobial property against pathogenic microorganisms. Soil samples collected from Kanyakumari district recorded the highest number of active actinomycetes followed by Coimbatore and Tirunelveli districts. The most active five actinomycetes were identified and characterized. These can be probed further for their potential application to control pathogens.
The authors are grateful to Vice Deanship of Research Chairs, King Saud University, Saudi Arabia for financial assistance.
The author declares no conflict of interest.

©2016 Devadass, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.