Journal of
eISSN: 2469 - 2786


Commonly used microbial site-specific recombinases used as editing and gene therapy tools in eukaryotic genomes suffer the need to pre-insert their recombination sites, or else need to undergo protein evolution to recognize endogenic pseudo sites. These difficulties can be bypassed using the lambdoid coliphage HK022 site-specific integrase recombinase that owing to its smaller integration site and sequence promiscuity can utilize active endogenous target sites.
Keywords: site-specific recombination, gene-editing, RMCE, gene therapy
bp, base pairs; CRISPR, clustered regularly interspaced short palindromic repeats; DSB, double-stranded dna break; GOI, gene of interest; gRNA, guide RNA; IHF, integration host factor; O, overlap; RMCE, recombinase-mediated cassette exchange; RS, recombination site; TALEN, transcription activator-like effectors; Xis, excisionase; ZFN, zinc finger nuclease
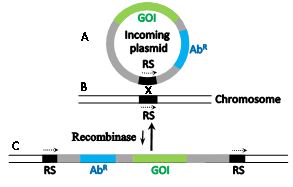
Site-specific recombinases catalyze recombination reactions between DNA pairs each carrying an identical but specific short sequence known as site-specific recombination site (RS). They differ in this regard from homologues recombinases that recombine practically any pair of homologous DNA sequences usually much longer than the RSs. Early attempts to use native homologous recombinases to accomplish eukaryotic gene manipulations proved inefficient. However, microbial site-specific recombination systems proved more useful for these purposes, initially when Brian Sauer introduced the Cre-lox system of Bacteriophage P1 into mammalian cells1 which was followed by the implementation of yeast's Flp-frt system. The RSs of these respective recombinases (lox and frt, respectively) are each 34 base pairs (bp) long and each comprises two complete or partial inverted repeats of 13 bp (Figure 1a, b, capital letters)that flank a central polar sequence of 8 bp (Figure 1a, b, small letters), known as the overlap (O). The sequences that include the inverted repeats (blue arrows in Figure 1) serve as binding sites for the relevant recombinase and O is the site of the DNA exchange reaction. Such RSs are usually absent in the genomes of higher eukaryotes. Therefore, Cre-lox or Flp-frt mediated genetic manipulations necessitate the introduction of the suitable RSs at specific genomic sites. Subsequently, a plasmid-borne gene of interest (GOI) abutted by a second compatible RS and a selection marker (usually an antibiotic resistant gene, Figure 2A) is used to selectively insert the GOI into the genome-inserted RS (Figure 2B) in a reaction catalyzed by the relevant recombinase that is provided in trans. As a result the GOI is site-specifically inserted along with the superfluous selection marker and extra plasmid DNA (Figure 2C). A major disadvantage of this procedure is that the inserted plasmid, flanked with two tandemly-oriented RSs (Figure 2C, broken arrows), serves as a substrate for the thermodynamically favorable reverse excision reaction (long vertical arrow in Figure 2) probably because of their cis configuration (both relatively adjacent on the same DNA molecule). Hence, it was easier to create gene knockouts when two RSs were inserted in tandem on each site of the gene to be excised.2,3 Another popular microbial site specific gene editing tool is the Integrase system (Int-att) of the Streptomyces phage fC31. In contrast to Cre-lox and Flp-frtthe two attRSs of fc31-Int (attB and attP) are not completely identical, 34 and 39 prospectively (Figure 1c).4 attP x attB recombination catalyzed by its integrase recombinase (Int-fC31) results in the recombinantattL x attR sites that flank the insertion; however, in the mammalian milieu, they are not reversible thereby allowing stable integrati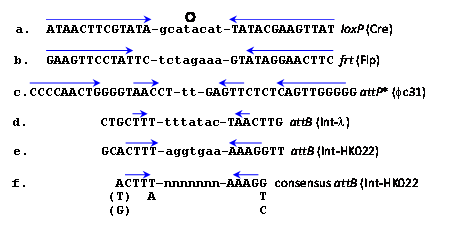 on. Moreover, mammalian genomes carry some partially mismatched but active native attP sites (pseudo attPsecondary sites) that were used as successful gene editing and gene therapy targets.5
on. Moreover, mammalian genomes carry some partially mismatched but active native attP sites (pseudo attPsecondary sites) that were used as successful gene editing and gene therapy targets.5
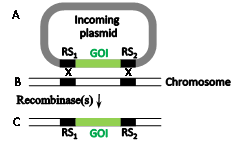
The insertion problem of entire plasmids was partially solved with the implementation of a dual site-specific recombination reaction known as the "recombinase-mediated cassette exchange" (RMCE) reaction (Figure 3), first implemented in Jürgen Bode's laboratory using the Flp-frt system.6 Here, a chromosomal sequence flanked by two incompatible RSs (RS1 and RS2, Figure 3B) is exchanged with a plasmid-borne sequence (Figure 3A) likewise flanked by RS1 and RS2 (Figure 3C). It is essential the RS1 and RS2be incompatible to avoid recombination between them. If only one recombinase catalyzes the RMCE reaction it must recognize both compatible recombining RS pairs (at least one pair is mutated). Alternatively each compatible RS pair can match a different recombinase, in which case both recombinases must be supplied.2,7 A Cre-catalyzed RMCE reaction that swapped fragments over 100 kb long was accomplished in mice.8 The need to pre-insert Cre and Flp RSs (loxP and frt, respectively) was partially overcome by the search and use of native secondary RSs with some mismatches that are inactive with their wild type recombinases. Rendering a recombinase specific for such secondary sites entails a lengthy directed protein evolution process. Several such modified recombinases able to mediate gene-editing reactions with endogenous secondary RSs were created in this manner.9-14 Several other microbial site-specific recombinases that are less popular in eukaryotic gene manipulations are listed in.15
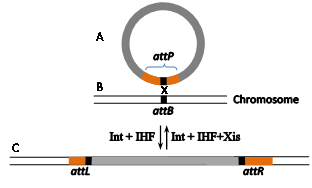
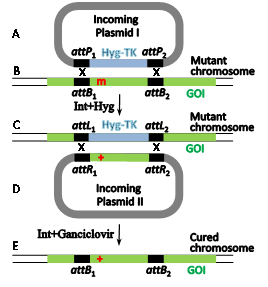
The well-documented site-specific recombination system Int-att of the temperate coli phage l (Figure 4) has been much less employed as a gene editing tool. Its bacterial attB RS is considerably shorter (21 bp, Figure 1d, Figure 4B). It comprises two partially inverted 7 bp Int-binding sites flanking a 7bp O region. However, the reason for its lingering behind is that attB's21bp phage partner RS (attP) is flanked by two longer arms (188 and 74bp, brown rectangles in Figure 4A) that carry additional indispensable binding sites forint as well as binding sites for the indispensable host and phage-encoded accessory proteins IHF and Xis.16,17 As with fC31 RSs, an attB x attP integration reaction leads to the formation of flanking recombinant attL and attR sitesof the inserted DNA (and the prophage in nature, Figure 4C). A similar site-specific recombination mechanism characterizes the l-related coliphage HK022.18,19 While both lambdoid Int systems were recruited to function as gene editing tools in mammalian cells, in contrast to the Int-att system of fC31, their reverse attL x attR excision reactions are also functional in mammalian cells. However, only Int-HK022 is able to catalyze site-specific recombination reactions in the absence of the accessory proteins, albeit, at reduced efficiency.20,21 For l to do so, an IHF-independent mutation of Int is used.22 The freedom from the accessory proteins rendered unnecessary the cognate attP¬-HK022arms.23 Moreover, it turned out that replacing the O sequence of attB-HK022with a random 7bp sequence still supports efficient Int-mediated site-specific recombination as long as its cognate phage recombination site attP features an identical O sequence.24 This property, combined with the relative shortness of attB allowed identifying native and active secondary attB sites that flank several human deleterious mutations, each with a different O sequence, and some even with minor modifications in their Int-binding sites. Owing to the difference in their O sequences renders the flanking secondary attBs incompatible with each other. Hence, such active endogenous sites that flank human hereditary mutations (Figure 5B) are potential RSs in RMCE reactions catalyzed by unmodified Int-HK022 thereby super seeding the need to insert any attB sites or alter the specificity of the recombinase by directed protein evolution. Some of these flanking attBs are currently exploited for performing two sequential RMCE reactions catalyzed by wild type Int-HK022 with the aim cure such mutations (Figure 5).The reason of the two RMCE reactions is for selection purposes and for the recovery of the native attB s that leads to “cleanly” cured cells that, except of the cured mutation, are devoid of any other genome DNA modification (Figure 5E).
An analysis of several potential human native secondary attB-HK022 sites that were either active or inactive, along with minor mismatches in the Int binding sites that flank a random O sequence lead to a consensus sequence of a 15 bp (with some flexibility) active attB-HK022 site that maintained its CTT….AAG inverted repeat (Figure 1f).24 In l, minor changes in its O sequence likewise did not affect recombination provided identical in both recombining att partners.25,26 Whether in l’s att sites as well as in loxP and frt an extended randomness in the O sequence of their recombining RS pairs allows recombination was not yet reported.
Recent gene editing approaches based on the recruitment of the endogenous homologous recombination repair system in response to double-stranded DNA break (DSB) seem to supplant those based on the microbial site-specific recombination systems. Key to the success of the homologous recombination repair systems is the ability to introduce DSBs at any desired chromosomal loci. This feat has been originally accomplished using sequence-specific DNA-bound Zinc-finger double-stranded DNA nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) fused to a blunt-end restriction enzyme motif (usually FokI). More recently, these engineered DNA-bound nuclease proteins are replaced by site-specific DSB induction mediated by engineered derivatives of the bacterial adaptive immunity CRISPR/Cassystem.27,28 However, to accomplish productive RMCE reactions these systems still associate with site-specific recombinases. E.g. the CRISPR/Cas system was already used to insert and perform RMCE reactions in Drosophila based on the activity of Int-fC31 system29 and a seed-mouse line was constructed that uses the Cre-lox based RMCE reactions in the mouse Rosa26 locus.30,31 Namely, these RMCE constructs still use pre-inserted RSs of the prokaryotic site-specific recombinases and they are not necessarily located in their native genome location. The proposed int-HK022 RMCE reactions, where possible, have the spatial advantage of using native RS sequences to cleanly correct deleterious mutations within their native loci (Figure 5).
Gabi Kaufmann’s valuable comments and corrections helped improved the manuscript. Work done in our laboratory was supported by GIF, the German Israeli Foundation for Scientific Research and Development (Grant 1062/2008) the Israel Science Foundation (Grant 702/11) and the US-Israel Binational Science Foundation Jerusalem, Israel (Grant 2003394).
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.