Journal of
eISSN: 2473-0831


Research Article Volume 10 Issue 3
1Nigerian Institute for Trypanosomiasis & Onchocerciasis Research, Ibadan, Nigeria
2 Department of Biochemistry, Faculty of Pure and Applied Sciences, Federal University Wukari, Nigeria
3Waterspring International School, Nigeria
Correspondence: Akinseye Olanrewaju Roland, Nigerian Institute for Trypanosomiasis & Onchocerciasis Research, Ibadan, Nigeria, Tel +2347031828 325
Received: May 14, 2021 | Published: June 9, 2021
Citation: Roland AO, Morayo AE, Joan O, et al. Modelling profile of onchocerca volvulus glutamate-cysteine ligase (ONCVO-GCL). J Anal Pharm Res. 2021;10(3):118-122. DOI: 10.15406/japlr.2021.10.00374
Onchocerca volvulus Glutamate cysteine ligase (ONCVO-GCL) catalyzes the first step in the production of the cellular antioxidant glutathione (GSH), which involve the condensation of cysteine and glutamate to form the dipeptide gamma-glutamylcysteine (γ-GC). ONCVO-GCL is critical to cell survival. Its dysregulation could lead to decreased GSH biosynthesis, reduced cellular antioxidant capacity, and the induction of oxidative stress. ONCVO-GCL expression support the high level of cell proliferation and confer resistance to many chemotherapeutic agents, hence could serve as a molecular target for inhibitors. This study aims to model the 3-dimensional (3D) structure of ONCVO-GCL, validate and predict the active sites of the modelled protein. ONCVO-GCL (Uniprot ID: A0A044QR48) 3D structure was modelled and validated using SWISS-MODEL. The Computed Atlas of Surface Topography of proteins (CASTp) 3.0 was used to predict the active sites of the modelled protein. A percentage identity matrix of 41.81% was obtained, which confirms the similarity identity of 40.86% obtained from the homology modelling. Model with 88% in the most favoured region of Ramachandra plot was obtained and the more favourable active sites for docking analyses due to the similarities observed from the alignment of the modelled structure to the template structure were: GLY 2A, LEU3A, LEU 4A, ARG 40A, TRP 47A, GLY 48A, ASP 49A, GLU 50A, GLU 52A, and PRO 109A.
Keywords: glutamate cysteine ligase, modelling, active site, target
Onchocerca volvulus, a causative agent of river blindness in human has been reported to have infected in 2017, an estimation of 20.9 million people worldwide, causing severe eye damage and eventual blindness. Glutamate cysteine ligase (GCL) is also referred to as gamma-glutamylcysteine synthetase (GCS). It is the starting enzyme as well as the rate-limiting enzyme in the pathway for biosynthesis of glutathione (GSH) in the cell as indicated in the reaction;
L-glutamate + L-cysteine + ATP 🠦 gamma-glutamyl cysteine + ADP + Pi
Onchocerca volvulus Glutamate cysteine ligase (ONCVO-GCL) catalyzes the ligation of cysteine and glutamate yielding gamma-glutamylcysteine (γ-GC), a dipeptide. ONCVO-GCL is critical to cell survival which is essential for GSH in this organism and other filarial worms.
The dysregulation of this enzyme has been reported nearly in all eukaryotes in which the expression of GCL protein is found, including plants and humans. Genetic knockdown of GCL has shown to results in embryonic lethality,18 as well as most diseases of human, including diabetes.19,20 Consequent to the strong immune system of the host, parasites (nematodes) experience intensified oxidative stress,1 but GSH, an antioxidant offers a protection. Hence, the levels of reduced GSH must be maintained at a substantial amount for the survival of the parasite.
The GSH levels in the cell as well as its biosynthetic capacity is often dependent on the activity of glutamate-cysteine ligase.21,22,23 As a result of its function as the enzyme in the rate-limiting step during glutathione biological synthesis, differences in the activity of GCL directly proportional to the differences in the glutathione cellular synthesis.20 In the light of this, efforts in altering the production of GSH have always been targeted towards this enzyme.24
In the past years, scientist have adopted several clinical techniques in the design and production of therapeutic agents in commercial quantity as drugs.2 One major method is the Structure-based drug design (SBDD) makes use of the 3-dimensional (3D) structure of the target protein to produce appropriate ligand that can bind it and serves as an inhibitor.3 In a situation where there is no experimental information about the 3D structure of the protein, its amino acid sequence is consulted to design a homology model.3,25 This study therefore aims at modelling, designing the 3D structure of ONCVO-GCL and predicting the active site of the protein.
Homology modelling of ONCVO-GCL and the target template sequence alignment
The crystal structure of ONCVO-GCL is absent in the Protein Data Bank (PDB);4 its 3D structure was therefore modelled. The ID of the protein target (Onchocerca volvulus Glutamate-Cysteine Ligase-1) was obtained from UniProt Knowledgebase (UniProtKB) (UniProt Consortium, 2018) with the accession number A0A044QR48, after which the protein ID was submitted to SWISS-MODEL web server,6 to design a model with sufficient query sequence coverage and sequence identity. Global Model Quality Estimation (GMQE) was consulted in choosing the best 3D structure,7 and Qualitative Model Energy Analysis (QMEAN),8 values. The values of GMQE are usually between 0 and 1, the reliability of the predicted structure is dependent on the number. In case of QMEAN, reliability is indicated by the value below 4.0.9,25 Clustal Omega version 1.2.1 was used to confirm the similarity between the amino acid sequences of the homology model of ONCVO-GCL and the template structure used for the homology.10
Modelled protein structure validation
The QMEAN scoring function for estimating the quality of the local and the global model was calculated automatically based on the geometry, the interaction and the protein model solvent potential by the SWISS-MODEL web server. It also gives the z-score of 0 to 1 range, to which the expected value for any structure are compared.25 The quality of the modelled 3D structure of ONCVO-GCL produced through SWISS-MODEL was examined by PROCHECK. The modelled ONCVO-GCL pdb file format was uploaded on the PDB sum web server of European Bioinformatic Institute for this structure validation.11 The Ramachandran plot and the Ramachandran plot statistics were gotten by uploading the modelled ONCVO-GCL pdb file format on the server. The information about the amount of amino acid residues located in the favourable, allowed and disallowed regions is given by the Ramachandran plot statistics,12,25 though the assessment of the quality of a modelled protein or an experimental structure is carried by the Ramachandran plot. Also the modelled protein structure validation as well as its compatibility to its own amino acids were ascertained by Verify3D.13 This also compared the structure of the result with the well-known structure.
ONCVO-GCL model alignment and the template structure
The ONCVO-GCL model alignment and structure of the template was carried out with the aid of PyMOL molecular viewer,14 to reveal the level of relationship among the carbon atoms. This is obtained from the root mean square deviation (RMSD) between the carbon atoms positioning of both the model and the template which is got from the alignment.25 The degree of relationship between the structures depends on how low the RMSD (w.r.t 0) is.25
align 3ig5_Template*, Verify3D_OvGCL-1_model.pdb*, cycles=0, transform=0
Active sites determination in the modelled protein
The active sites in the modelled protein structure was predicted using the Computed Atlas of Surface Topography of proteins (CASTp) 3.0.15 The identification and measurement of voids on 3D protein structures was carried out with CASTp, an online server.16 The modelled 3D protein was submitted on the server, and the necessary amino acids for binding interactions were predicted.16
Homology modelling of ONCVO-GCL and the target template sequence alignment
SWISS-MODEL with GMQE of 0.74 and QMEAN of -3.08 was used in building a 3D structure of ONCVO-GCL. Also, Saccharomyces cerevisiae glutamate cysteine ligase in complex with Mg2+ and L-glutamate (PDB ID: 3IG5; resolution: 2.10 Å) was identified to have the closest template to ONCVO-GCL with a similarity identity of 40.83% and sequence similarity of 0.40. The GMQE value of 0.74 and QMEAN score of -3.08 show that the modelled structure is reliable and has a good quality.7,9
Figure 1 shows the multiple sequence alignment of the amino acid sequences,17 of the ONCVO-GCL (UniProtKB ID: A0A044QR48) and S. cerevisiae GCL in bound with Mg2+ and L-glutamate (PDB ID: 3IG5). A percentage identity matrix of 41.81% confirming the similarity in the identity of 40.83% obtained from the homology modelling was obtained.

Figure 1 Alignment of the amino acid sequences of Onchocerca volvulus GCL and the crystal structure of 3IG5.
‘*’represents positions that have single, fully conserved residue; ‘:’ indicates conservation between groups of strongly similar properties; ‘.’ indicates conservation between groups of weakly similar properties.
Abbreviation: GCL, glutamate cysteine ligase.
Structure validation of modelled protein
The plot of the predicted local similarity was represented graphically to target against the residue number of the predicted modelled protein 3D structure Figure 2(A).25 Majority of the residues value was close to 1, depicting that estimate of the local quality of the residues of the predicted model is good. The residues having values below 0.6 were deemed to be low in quality. The modelled protein designed structure are also formed within the distance of other protein structures in PDB, which ascertains its reliability Figure 2(B).
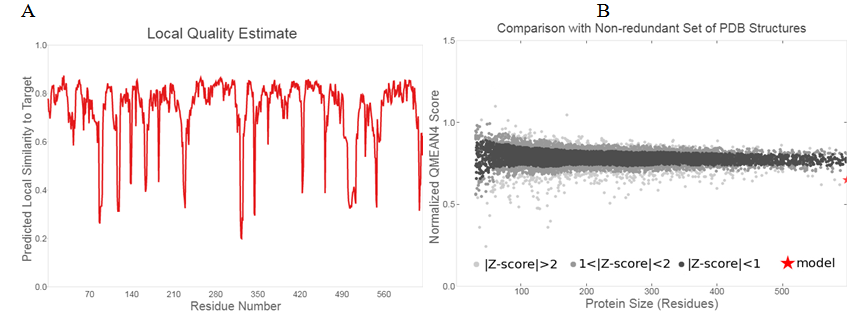
Figure 2 Structure validation of modelled ONCVO-GCL: (A) Local quality estimate of the residues of the predicted ONCVO-GCL model; (B) comparison of the predicted ONCVO-GCL structure with non redundant set of PDB structures.
Abbreviations: ONCVO-GCL, onchocerca volvulus glutamate cysteine ligase; PDB, protein data bank.
The Ramachandran plot Figure 3(A) and the Ramachandran plot statistics Figure 3(B) were both obtained from web server of PDBsum. The Ramachandran plot statistics indicated that the modelled 3D structure of ONCVO-GCL has 88% of its residues in the most favoured regions, 11.2% of its residues in additional allowed regions, 0.7% of its residues in the generously allowed regions and 0.0% of its residues in disallowed regions of the Ramachandran plot. The structure of modelled 3D protein is also validated by this to be a model of good quality. Also, the Verified 3D plot of the modelled protein showed as PASS Figure 3(C) when obtained for the structure validation. The environment profile of the 3D reveals that 90.24% of the residues have average 3D-1D score ≥ 0.2, which suggests that the modelled protein is valid.
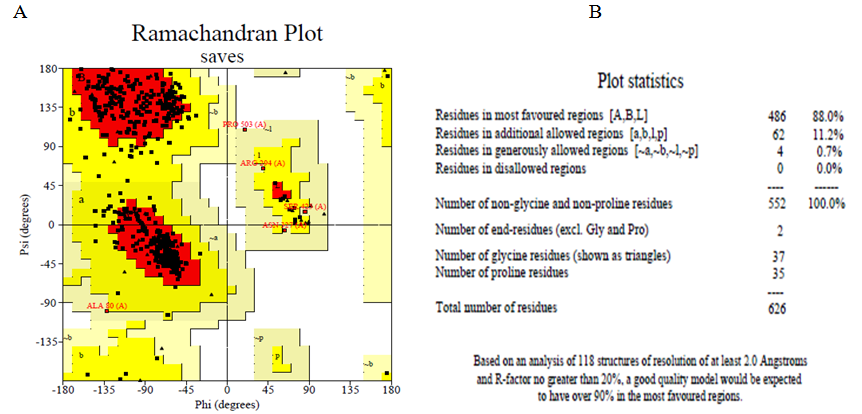
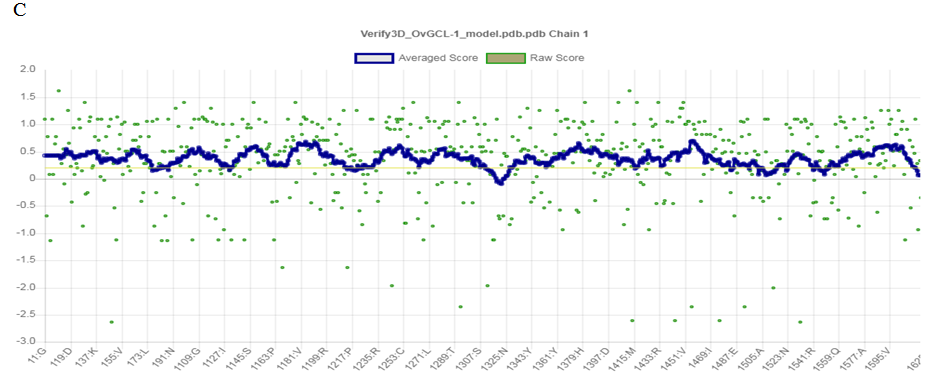
Figure 3 Structure validation using (A) Ramachandran plot; (B) Ramachandran plot statistics of the homology modelled ONCVOGCL; and (C) Verify3D.
Abbreviation: ONCVO-GCL, onchocerca volvulus glutamate cysteine ligase.
ONCVO-GCL model alignment and template (3IG5) structure
PyMOL molecular viewer was used to compute alignment and it gave a RMSD value of (3.728 Å) which indicates that the structures were slightly closely related (Figure 4). The ash colour helices represent the template structure, while the green helices represents the protein model. As shown by the alignment, the template structure (3IG5) chain A corresponded to the chain A of the protein model structure. More so, it was revealed by the molecular viewer that the Mg2+ binding to the amino acid residues of 3IG5 at GLU 96A, GLU 103A, GLU 52A and GLU 693A was also corresponding to the Mg2+ of the amino acid residues of the modelled template at GLU 106A, GLU 99A and GLU 52A. In 3IG5 template, there are GLU binding site which is not conserved and PGE sites which are not biologically relevant, which could not be found in the new modelled protein.
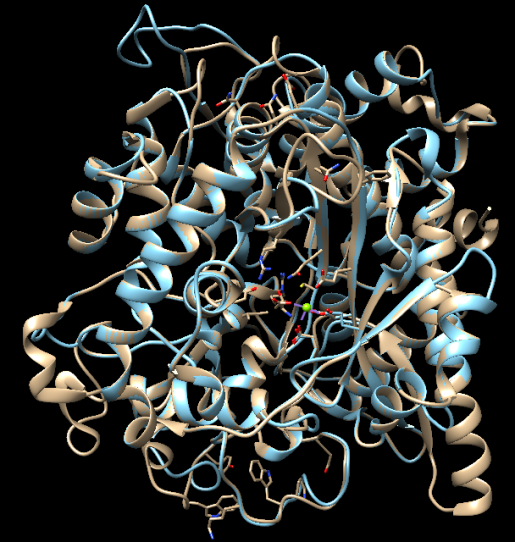
Figure 4 Alignment of the ONCVO-GCL model and the 3IG5 template structure of Saccharomyces cerevisiae GCL.
Abbreviation: ONCVO-GCL, onchocerca volvulus glutamate cysteine ligase.
Identification of active site
As predicted by the active site, a pocket of an area (SA) of 1123.097 and a volume (SA) of 1283.631 was identified (Figure 5). The modelled protein was predicted to have a total of 236 amino acid residue at the active site. Moreover, the more favourable sites were selected for the docking analyses base on the similarities found in the structure alignment of the modelled protein to the template: GLY 2A, LEU 3A, LEU 4A, ARG 40A, TRP 47A, GLY 48A, ASP 49A, GLU 50A, GLU 52A and PRO 109A.
Considering the fact that ONCVO-GCL is a significant drug target, it can employed in the design of inhibitors to tackle onchocerciasis. This study sheds light on the design as well as the prediction of potential active sites of homology modelled ONCVO-GCL. The model designed is 88% in the most favoured region of Ramachandra plot. Also, there is need for experimental based validation of the protein target.
None.
The author declares there is no conflict of interest.

©2021 Roland, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.