Journal of
eISSN: 2473-0831


Research Article Volume 10 Issue 2
1Research-Development Department, Center for Genetic Engineering and Biotechnology of Sancti Spiritus, Cuba
2Quality Control Direction, Center for Genetic Engineering and Biotechnology, Cuba
Correspondence: Maylin Pérez-Bernal, Center for Genetic Engineering and Biotechnology of Sancti Spiritus, Cuba
Received: December 20, 2020 | Published: March 8, 2021
Citation: Pérez-Bernal M, Hernández C, Delgado M, et al. ELISA validation approach for the detection of anti-Saccharomyces cerevisiae antibodies in patients treated with biopharmaceutical Heberprot-P®. J Anal Pharm Res. 2021;10(2):50-56. DOI: 10.15406/japlr.2021.10.00365
This work describes the validation of an enzyme-linked immunosorbent assay (ELISA) for detection of anti-Saccharomyces cerevisiae antibodies (ASCA) in diabetic patients with foot ulcers, after the treatment with Heberprot-P®. Validation followed regulatory guidelines of US FDA and European Medicine Agency. Minimum required dilution of samples and quality controls were defined using pools of sera from diabetic patients and from healthy donors. Parameters such as cut point, specificity, precision, selectivity, robustness and sample stability were analyzed. The repeatability and intermediate precision percent ranged between 7.93-10.61% and 7.93-11.43 %, respectively, indicating low intra- and inter-assay variation. The specificity was proved by background noise suppression, reaching 100% of inhibition as strong criterion for the specificity of the immunoassay. The validated ELISA is a reliable tool for ASCA detection in human serum after the administration of Heberprot-P®, in order to find immunological reactions associated with latent contamination by host cell proteins from Saccharomyces cerevisiae.
Keywords: anti-Saccharomyces cerevisiae antibodies, diabetic patients, ELISA validation, serum samples
ELISA, enzyme-linked immunosorbent assay; ASCA, anti-Saccharomyces cerevisiae antibodies
Saccharomyces cerevisiae is the most widely used yeast for recombinant protein production. This single-cell eukaryotic organism possesses the advantages of bacteria and eukaryotes: it is easy to culture, grows fast, can give high productivity,1 can ensure proper protein folding and post-translational modifications2 and it can secrete the product to the extracellular medium which simplifies purification.3 Moreover, as a generally recognized as safe organism, free of pyrogens also makes S. cerevisiae a favorable expression system for biopharmaceuticals.4 Even though S. cerevisiae is a good platform for biopharmaceuticals production, during the manufacture of such products, host cell-derived material will inevitably be introduced into the process stream.5 Such contamination can result in undesirable immunological reactions, by generating anti-Saccharomyces cerevisiae antibodies (ASCA) in patients that have been treated with the biopharmaceutical. ASCA including immunoglobulin IgG and IgA, which appear to be specifically directed against mannose sequences of mannan present in the cell wall of S. cerevisiae.6 At present, ASCA is one of the most commonly used serologic antibody markers for diagnosis of inflammatory bowel diseases.6,7 In addition, S. cerevisiae is common yeast found in various foods; consequently ASCA can appear in healthy persons too. For that, it is very important to know the serum levels of ASCA in the patients, pre- and post-treatment with any biopharmaceutical obtained from S. cerevisiae expression system, in order to recognize the ASCA associated with the bio-product.
Enzyme-linked immunosorbent assay (ELISA) is frequently utilized for immunogenicity recognition. The current available ELISA methods for the detection of ASCA agree well but differ greatly in the interpretation of results. It is clear that cut points have been chosen for different purposes, lipemic and hemolytic samples should not be used or have been used with cautions, and the thermostability of the samples is not the same in different ASCA assays.8 In addition, commercial ELISA kits are expensive and not always readily available. Particularly for the clinical studies related with Heberprot-P®, a biopharmaceutical produced by S. cerevisiae, with direct effects on granulation and epithelialization of diabetic foot ulcers, it's still necessary the validation of an ELISA for the intended purpose of ASCA detection in human serum after the administration of this biopharmaceutical, as a reliable method to detect immunological reactions associated with latent contamination by host cell proteins from yeast. The aim of this work was to validate an ELISA for the detection of ASCA in serum from diabetic patients with foot ulcers after the treatment with Heberprot-P®. Validation parameters suggested by regulatory guidelines of US Food and Drug Administration9,10 and European Medicine Agency11 were assessed. As the percent of inhibition to validate the specificity has not been specified for guidelines, this work included a method to eliminate the possible background noise that can affect the percent of inhibition, in order to consider a percent close to 100% as a strong criterion that supports the specificity of the immunoassay.
Origin of human serum
Individual human serum samples obtained from 30 healthy donors and pools of sera from 30 diabetic patients with foot ulcer, before and after the treatment with Heberprot-P®, were used for the validation protocol. All sera were stored at −20°C until use.
Enzyme-linked immunosorbent assay
Polystyrene 96-well microtiter plates (Costar™ High Binding 3590), were coated with 10 µg/mL of the antigen (Reference Material of contaminants from S. cerevisiae as host cell for Recombinant Epidermal Growth Factor, lot 03SFD16002N, CIGB, Havana, Cuba) in 100 μL/well coating buffer (phosphate-buffered saline, pH 7.2) and incubated 1h at 37°C. Wells were washed four times with 350 μL/well washing buffer (0.292 M Tris (hydroxymethyl) aminomethane, 0.228 M HCl (37%), 3.75 M NaCl and 1.25 % Tween-20, pH 8.0) and blocked with 350 μL/well blocking buffer (washing buffer with 1% nonfat dried milk powder, pH 8.0) 1h at 37ºC. The plate was washed once with washing buffer and 100 μL of quality controls and serum samples diluted with blocking buffer were added to wells and incubated 1h at 37ºC. Plates were washed four times and 100 μL anti-human polyvalent alkaline-phosphatase-conjugated (Sigma; 1:5000 in blocking buffer) was added to each well and incubated (1h, 22-25ºC). After four washes, 100 μL/well 1.0 mg/mL p-nitrophenyl phosphate, 1.0 M di-ethanolamine and 0.5 mM MgCl2, pH 9.8, was added and incubated 30 min at 28ºC in the dark. Color development was stopped by adding 100 μL/well of 0.1 M di-sodium EDTA. Absorbance was measured at 405 nm (A405nm) in a microplate reader (Labsystems Multiskan® Plus, Finland).
Minimum required dilution (MRD)
The MRD was determined as the minimum dilution that can maximize the difference between the A405nm of a pool of sera from diabetic patients with foot ulcer before treatment with Heberprot-P®, and the A405nm of a pool of sera from healthy donors, with the minimum photometric error (A405nm close to 0.432). Both pools of sera were two-fold serial diluted, from 1:1600 to 1:100. Each dilution point was duplicated in the ELISA plate.
Quality controls
Three quality controls (QCs) were prepared with a pool of sera from healthy donors diluted with blocking buffer as follows:
Negative control (NC): Pool of sera diluted at MRD and pre-incubated with 100 µg/mL of the antigen used for coating, 1h at (22-25) ºC.
High Positive Control (HPC): Pool of sera diluted 1:20
Low Positive Control (LPC): Pool of sera diluted 1:75.
Once the QCs were prepared, the next step was to determine the range of acceptable values for them. QCs were studied in triplicates by two analysts in six independent runs, over three different days each one. Outliers were identified by the Box Plot method and removed from the runs. The NC complied the acceptance criterium if it not above the one-sided confidence interval (99%) determined by the formula: mean (logA405nm) + t (0.01; DF) x SD (standard deviation). HPC and LPC should be in their two-sided confidence intervals (99%), calculated by using the ratio PC/NC, established between A405nm of corresponding positive control and A405nm of negative control. The intervals were determined by the formula: mean (log PC/NC) ± t (0.01; DF) x SD.
Cut-point
The cut-point (CP) was defined by analyzing 34 individual human serum samples from diabetic patients with foot ulcer before treatment with Heberprot-P®. Samples were diluted at MRD and studied in duplicates by two analysts in six independent runs, over three different days each one. A parametric approach (one sided, 99% confidence level) was selected to calculate the cut-point. The variable was the log-transformed S/N. The ratio S/N (signal/noise) was established between the A405nm from serum samples and the A405nm from negative control. Outliers were identified by the Box Plot method and removed from the runs. Data normality was analyzed using the Shapiro–Wilk test and the variance homogeneity was evaluated by the Levene test. A single factor ANOVA was applied to evaluate if the means of the runs were significantly different or not. All hypothesis tests were achieved with α=0.05. SD was estimated by performing a variance component analysis using restricted maximum likelihood method within the framework of a nested ANOVA. The cut point was calculated by the formula:
where 2.33 is the 99th percentile of the normal distribution
Specificity
The evaluation of specificity was performed by an inhibition assay. A pool of sera from 20 diabetic patients with foot ulcer after the treatment with Heberprot-P®, diluted at MRD, was pre-incubated with 100 µg/mL of the antigen during 1h at (22-25) ºC. The non-inhibited sample had PBS instead of the antigen. The inhibited and non-inhibited specimens were placed in triplicates in the ELISA plates, using two variants with the aim of background noise suppression: plate coated with the antigen and blocked as described before, and plate without coating but blocked. The percent of inhibition (%I) was calculated as follows:
%I = 100 – [(A405nm inhibited sample) / (A405nm non-inhibited sample)] x 100
ELISA was considered specific if the mean of the percent of inhibition was close to 100 %.
Precision
Precision was evaluated by repeating the assay in six analytical runs by two analysts, using as samples the negative control and three pools of sera with high, medium and low concentration of ASCA, obtained from healthy donors. The samples with high and medium concentration of ASCA coincided with the HPC and LPC, respectively. But the sample with low concentration of ASCA was prepared by diluting the pool of healthy donors 1:500 with blocking buffer. Each sample was evaluated in triplicates. The ratio S/N in logarithmic scale was used as dependent variable. SD was estimated by a variance component analysis using restricted maximum likelihood method within the framework of a nested ANOVA. Intra- and inter-assays precision, expressed as percent of coefficient of variation (%CV) at each concentration level, should not exceed 20%, and was calculated according to the formulas:
Selectivity
Selectivity was evaluated by testing two types of samples: eight individual hemolytic sera and eight individual lipemic sera, diluted at MRD. Serial two-fold dilutions, from 1:20 to 1:2560, were performed in duplicates for each individual serum, for the HPC and for each serum spiked with the HPC. Absorbance of individual sera, HPC and spiked sera, were estimated at MRD by plotting A405nm versus dilution factor. The percent of recovery (%R) was calculated by the formula:
Sample was considered not interfering if the percent of recovery was between 80 and 120%.
Robustness
There were made small changes in critical parameters of the ELISA procedure: variation in ±5 min the plate incubation times for antigen coating, for QCs and for anti-human polyvalent alkaline-phosphatase-conjugated; variation in ±0.2 mg/mL the substrate concentration. All treatments were assayed by triplicates. The robustness was proved by the fulfilling of the QCs confidence intervals.
Stability of samples and quality controls
Since the QCs and samples are of the same species, the stability of both was proved by the fulfilling of the QCs confidence intervals (99%). Short-term stability (6 hours at room temperature (20-25) ºC and 6 days at (2-8) ºC) and freeze–thaw stability (six freeze-thaw cycles), were tested using the QCs acceptance limits. For the evaluation of freeze–thaw stability, aliquots of each QC were thawed unassisted at room temperature. When completely thawed, the aliquots were refrozen at -20ºC for at least 20 hours. This freeze-thaw cycle was repeated five more times.
Statistical analysis
Statistical analyses were performed using Microsoft® Office Excel (2010) and the Statistical Package for Social Science 15.0. Curve fitting were achieved using Sigma Plot 12.
Minimum required dilution
The MRD is the dilution that yields a signal close to the signal of non-specific binding of assay diluent.12 High incidences of pre-existing ASCA are commonly found in healthy people sera, and it is difficult to obtain a suitable number of true negative samples and consequently to find an MRD ranged from 1:5 to 1:100 as recommended by FDA.9 Taking into account this inconvenience, in the present work the MRD was determined as the minimum dilution that maximizes the difference, with the minimal photometric error, between the A405nm of a pool of sera from diabetic patients, with foot ulcer before treatment with Heberprot-P®, and the A405nm of a pool of sera from healthy donors. Five 1:2 serial dilutions were performed in both pools of sera. Dilution factor versus A405nm were plotted using a logistic regression of five parameters (Figure 1). In the Figure 1A, a dilution factor of 500 was obtained by interpolating A405= 0.432 as the minimal photometric error. Then, this dilution factor was interpolated in the curve corresponding to the pool of healthy donors and it was obtained A405nm= 0.077. The ratio of both A405nm values was 5.6. Subsequently, this ratio was calculated using the A405nm of both pools obtained with 100 as dilution factor and the result was 4.1, a value 1.4 fold-low than the ratio obtained with 500 as dilution factor. This result proved the suitability of the use of 1:500 as MRD, because it maximized the difference between the A405nm of a pool of sera from diabetic patients with foot ulcer before treatment with Heberprot-P®, the target population, and the A405nm of a pool of sera from healthy donors.
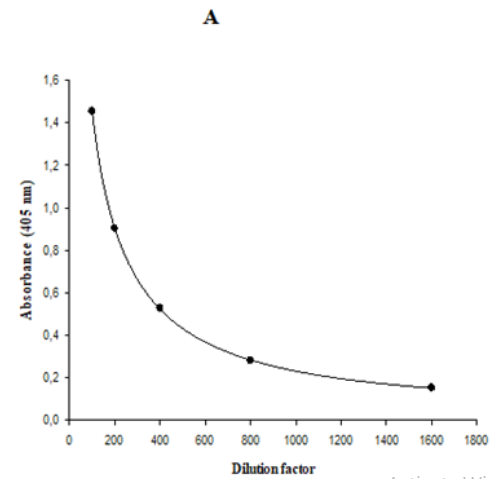
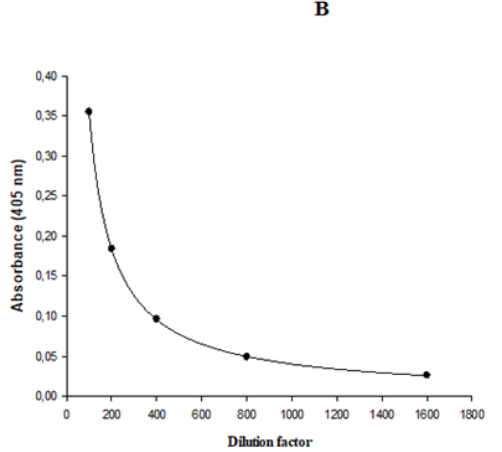
Figure 1 Representation of logistic regression of five parameters for 1:2 serial dilutions of the pool of sera from (A) diabetic patients with foot ulcer before treatment with Heberprot-P® and (B) healthy donors.
Quality controls confidence intervals
As S. cerevisiae is common yeast found in various foods, ASCA can appear in healthy persons and is very difficult to find a serum without ASCA as negative control. As novelty for this work is the use of a negative control, prepared with a pool of sera diluted with blocking buffer at MRD and pre-incubated with 100 µg/mL of the antigen used for plate coating. The purpose of obtaining 36 data points by running each QC was to quantify normal variation and establish the one-sided confidence interval (99%) for negative QC and the two-sided confidence intervals (99%) for low and high positive QCs (Table 1). Three outliers were identified by the Box Plot method and were not included when calculating the confidence intervals. These intervals will be used to monitor the performance and acceptability of the ELISA during subsequent validation parameters and clinical assays.
|
Quality |
One-sided confidence interval (99%) |
|||
|
Control |
log (mean A405nm) |
t (0.01; 10 DF) |
SD |
|
|
NC |
-1.669 |
2.764 |
0.052 |
0.03 |
|
Two-sided confidence interval (99%) |
||||
|
log (mean PC/NC) |
t (0.01; 11 DF) |
SD |
||
|
LPC |
1.537 |
3.106 |
0.056 |
low: 23.12; high: 51.22 |
|
HPC |
1.88 |
3.106 |
0.052 |
low: 52.24; high: 110.21 |
Table 1 Definition of the confidence intervals for the quality controls of the ELISA
NC, negative control; LPC, low positive control; HPC, high positive control
A405nm, Absorbance measured at 405 nm; SD, Standard deviation; DF, degrees of freedom
PC/NC, ratio A405nm of positive control/ A405nm of negative control
One-sided confidence interval (99%) for NC, mean (logA405nm) + t (0.01; DF) x SD
Two-sided confidence interval (99%) for LPC and HPC: mean (log PC/NC)±t (0.01; DF) x SD
Cut-point
In determining the CP, is important to use test samples as similar as possible to the study samples.12 In the present assay, the CP was established by analyzing individual human serum samples from diabetic patients with foot ulcer before treatment with Heberprot-P®, which are the study samples. The logarithm of the ratio S/N was used as variable; 42 outliers obtained from five consecutive iterations were removed, and 162 values of S/N remained for the determination of CP. The Shapiro–Wilk test (p≥0.05) demonstrated the normality of log S/N data. The p-value from the Levene test was 0.95, suggesting that the variances across assay runs were not significantly different at alpha=0.05 significance level. The single factor ANOVA demonstrated that means were also not significantly different (p=0.97) between runs. These results sustained the use of a fixed cut point for the assay. The media of log-transformed S/N was 0.45, and the standard deviation was calculated by restricted maximum likelihood method within the framework of a nested ANOVA. The components of variance were the 'analyst' and 'day' factors, but the factor 'error' was exclusively responsible for variance (Table 2). Substituting the mean value and the standard deviation into the formula log PC=media (log S/N) + 2.33xSD, a cut point of 15.57 was obtained.
|
Variance components |
Estimation |
|
Analyst |
0.00 |
|
Day (Analyst) |
0.00 |
|
Error |
0.10 |
|
Total Variance |
0.10 |
|
Standard Deviation |
0.32 |
Table 2 Estimation of standard deviation of log-transformed S/N by restricted maximum likelihood method within the framework of a nested ANOVA
S/N, Ratio (A405nm of each sample)/(A405nm of negative control)
Specificity
The specificity of an antibody refers to its ability to bind to the antigen of interest, but not to other assay components such as surfaces or reagents.9 A straightforward approach to addressing specificity is to demonstrate that binding can be blocked by soluble antigen in the inhibition step, as performed in the present study using a pool of sera from diabetic with foot ulcer before the treatment with Heberprot-P®. The possible background noise that can affect the percent of inhibition, was assessed by placing the inhibited and non-inhibited specimens in the ELISA plate coated with the antigen and blocked, and in the plate without coating but blocked. It was demonstrated that the method exhibited background noise affecting the specificity, possibly due to the blocking reagent,13 in which the non-specific inhibition percentage was 3.23% (Table 3). Background noise was suppressed by using the difference of signal obtained with coated and non-coated plates, to determine the percent of inhibition, which was close to 100% (Table 3). Taking into account the background suppression, the ELISA was considered specific.
|
Coated plate |
Non-coated plate |
Difference coated – Non-coated |
|
|
A405nm inhibited sample |
0.029 |
0.03 |
-0.001 |
|
A405nm non-inhibited sample |
0.287 |
0.031 |
0.256 |
|
% Inhibition |
89.9 |
3.23 |
100.39 |
Table 3 Evaluation of the specificity of the ELISA
Values of absorbance (A405 nm) represent the mean of three replications
%Inhibition =100 – [(A405nm inhibited sample) / (A405nm non-inhibited sample)] x 100
Some discrepancies are marked when assessing specificity for ligand binding assays. In FDA guidelines9,10 chapter of specificity is not given. Description related to specificity is contained in chapter of selectivity briefly and not in detail. The distinctions between the agencies concern sample concentrations, type of matrix, and concentration of interfering molecules,14 but the acceptance criterion for the percent of inhibition has been not specified for regulatory guidelines. The use of subjective criteria, such as ≥50% inhibition of signal, is discouraged.15 We consider that a percent of inhibition close to 100% is a strong criterion that supports the specificity of the immunoassay. To achieve this, it is necessary to guarantee the optimal conditions for the antigen-antibody reaction during the inhibition step, and to eliminate background noise as has been done in the present work.
Precision
The intra-assay precision (repeatability) expresses the precision under the same operating conditions over a short interval of time. The inter-assay precision (intermediate precision) validation verifies that in the same laboratory the method will provide the same results once the development phase is over.13 For our experimental conditions, the repeatability and intermediate CV precision percent ranged between 7.93-10.61% and 7.93-11.43 %, respectively (Table 4) indicating low intra- and inter-assay variation. These results indicated that the ELISA fulfills the specifications for precision, in accordance with guidelines for the validation of analytical procedures.9,11 The precision was sufficient to allow the direct comparison of samples processed by two analysts at different days.
|
Variance Components |
CV (%) |
||||
|
Sample |
Analyst |
Day (Analyst) |
Error |
Intra-assay |
Inter-assays |
|
NC |
0.000650 |
0.000131 |
0.004859 |
10.61 |
11.43 |
|
LCS |
0.000000 |
0.000595 |
0.00317 |
8.56 |
9.33 |
|
MCS |
0.000000 |
0.000000 |
0.003093 |
8.45 |
8.45 |
|
HCS |
0.000000 |
0.000000 |
0.002724 |
7.93 |
7.93 |
Table 4 Determination of precision intra- and inter-assays in six independent runs
NC, negative control; LCS, sample with low ASCA concentration; MCS, sample with medium ASCA concentration; HCS, sample with high ASCA concentration
S/N: Ratio (A405nm of each sample)/(A405nm of NC)
%CV intra-assay = [√ 10^ (variance of error) – 1] x 100
%CV inter-assays = [√ 10^ (√∑variance components) – 1] x 100
Selectivity
Selectivity is the ability of an analytical method to detect and differentiate the analyte in the presence of other components in the sample. In the present study, selectivity was assessed using two kinds of samples: hemolytic and lipemic sera, since the most common nonspecific interferences in ELISA assays are due to hemolysis and lipemia.16 According to the FDA9 and EMA11 guidelines, individual matrices for human should be spiked with a standard at or near the low limit of quantification to evaluate selectivity. In our experiments, a standard with known concentration of ASCA did not exist and it was necessary to apply a different method to evaluate the recovery in possible interfering samples. The method was based on serial dilutions of three types of specimens: individual serum samples (lipemic or hemolytic), the HPC and the mixture of both. Data of A405nm obtained from each specimen were plotted versus dilution factor using a logistic regression of five parameters. The value of A405nm at MRD was obtained by interpolation and it was used for calculating the percent of recovery in each probable interfering sample. The percent of recovery for each individual serum sample was in the acceptance range (Table 5), and it signified that the absorbance of the individual serum spiked with HPC was practically equivalent to the sum of the absorbances of individual serum and HPC, evaluated by separate. Thus, hemolysis and lipemia did not interfere in the capacity of the ELISA to detect the ASCA in serum samples.
|
A405nm obtained by interpolation at MRD |
||||
|
Code for individual serum samples |
Individual serum sample |
HPC |
Serum samples spiked with HPC |
%R |
|
H1 |
0.136 |
0.149 |
0.261 |
91.67 |
|
H2 |
0.158 |
0.196 |
0.321 |
90.5 |
|
H3 |
0.151 |
0.187 |
0.306 |
90.47 |
|
H4 |
0.146 |
0.16 |
0.278 |
90.6 |
|
H5 |
0.191 |
0.208 |
0.349 |
87.42 |
|
H6 |
0.17 |
0.21 |
0.342 |
89.83 |
|
H7 |
0.142 |
0.178 |
0.295 |
92.15 |
|
H8 |
0.162 |
0.2 |
0.327 |
90.49 |
|
L1 |
0.017 |
0.149 |
0.142 |
85.94 |
|
L2 |
0.173 |
0.196 |
0.329 |
88.69 |
|
L3 |
0.165 |
0.187 |
0.312 |
88.7 |
|
L4 |
0.022 |
0.16 |
0.154 |
84.34 |
|
L5 |
0.042 |
0.208 |
0.2 |
80.02 |
|
L6 |
0.186 |
0.21 |
0.348 |
87.93 |
|
L7 |
0.156 |
0.178 |
0.3 |
90.04 |
|
L8 |
0.176 |
0.2 |
0.333 |
88.56 |
Table 5 Evaluation of the selectivity of the ELISA by recovery estimation
H1 to H8, eight individual hemolytic serum samples; L1 to L8, eight individual lipemic serum samples; HPC, high positive control; %R, percent of recovery; MRD, minimal required dilution (1:500)
Robustness
The complexity of bioassays makes them particularly susceptible to variations in assay conditions, and it is essential to evaluate the robustness, assessed by the capacity of the assay to remain unaffected by small but deliberate variations in method parameters. For example, changes in temperature, incubation times, reagent lots and buffer characteristics.9,11 The robustness of this assay was evaluated by varying the incubation time in ±5min for antigen coating, QCs and anti-human polyvalent alkaline-phosphatase-conjugated; and by changing the substrate concentration in ± 0.2 mg/mL. All these variations did not alter the acceptability of QCs; the runs were accepted as “in-control” (Figure 2). This was a confirmation of the capability of the ELISA to remain unaffected by small deliberate changes in method parameters, and it provided an indication of its reliability during normal run conditions.
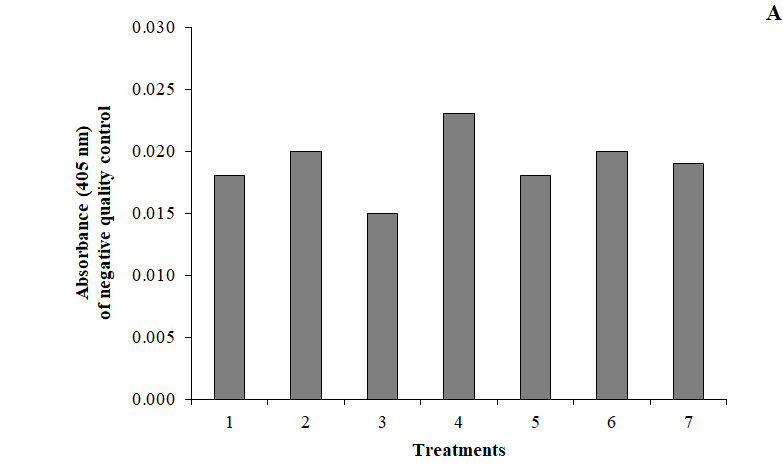
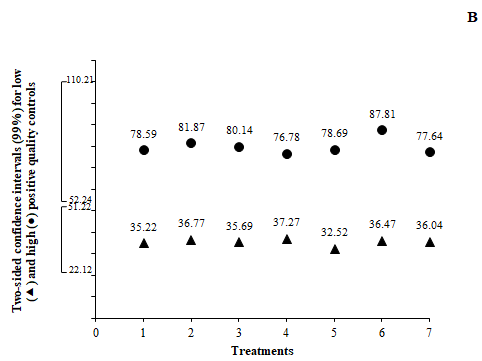
Figure 2 Evaluation of the robustness of the ELISA (treatments 1 to 4) and the stability of serum samples (treatments 5, 6 and 7) by the fulfilling of (A) negative and (B) positive quality controls confidence intervals. Data represents the mean of three replications. Treatments: (1) variation in -5 min the plate incubation times for antigen coating, for quality controls and for anti-human polyvalent alkaline-phosphatase-conjugated; (2) variation in +5 min the plate incubation times for antigen coating, for quality controls and for anti-human polyvalent alkaline-phosphatase-conjugated; (3) variation in -0.2 mg/mL the substrate concentration; (4) variation in +0.2 mg/mL the substrate concentration; (5) 6 hours at room temperature (20-25)ºC; (6) 6 days at (2-8)ºC; (7) 6 freeze-thaw cycles. Confidence intervals (99%) for quality controls:
A405nmNC≤0.030 22.12≤ LPC/NC≤51.22 52.24≤HPC/NC≤110.21.
Stability of samples and quality controls
Frozen serum material is usually stable for a long time, but its stability should be monitored over time because the assay lifetime can be very long.5 Determination of the number of freeze/thaw cycles, storage times and temperatures should be based on the expected handling and storage of the serum samples. If QCs have been prepared in the same matrix as the study samples, the use of QCs with high and low levels of antibody evaluates the stability of samples.11 In the present study, the negative and positive quality controls were applied for the stability assessment. For the evaluation of the stability, QCs were submitted at temperature conditions commonly used for the processing and storing of samples during the ELISA. The results of stability assessments confirmed that QCs were stable in all tested conditions, since all the measures were into the confidence intervals (99%) (Figure 2). The stability of QCs with high and low concentration of ASCA reflected the stability of the study samples, because the QCs and samples are of the same species.
An ELISA for the detection of ASCA in patients treated with Heberprot-P® was validated according to published guidelines. Positive and negative quality controls were prepared and their acceptance limits with 99% confidence were established to monitor the acceptability of the ELISA. As Saccharomyces cerevisiae is common yeast found in various foods, ASCA can appear in healthy persons and is very difficult to find a serum without ASCA as negative control. As novelty for this work is the use of a negative control, prepared with a pool of sera diluted with blocking buffer at MRD and pre-incubated with 100 µg/mL of the antigen used for plate coating. Since some discrepancies are marked when assessing specificity for ligand binding assays, the specificity was proved by a method of background noise suppression, reaching 100% of inhibition as strong criterion that supports the specificity of the immunoassay. The validation revealed that this ELISA is precise and robust to changes in incubation times and in substrate concentration. The stability of quality controls at short-term temperature and in freeze–thaw cycles was demonstrated, and it reflected the stability of the samples, since the quality controls and samples are of the same species. The validated ELISA is a reliable tool for ASCA detection in human serum after the administration of Heberprot-P®, in order to find immunological reactions associated with latent contamination by host cell proteins from Saccharomyces cerevisiae.
None.
The authors declare that they have no potential conflict of interests.
The Center for Genetic Engineering and Biotechnology (CIGB) is a Cuban institution of a dynamic development which has reached a high level of research and development of biopharmaceuticals (www.cigb.edu.cu).

©2021 Pérez-Bernal, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.