Journal of
eISSN: 2473-0831


Research Article Volume 8 Issue 2
1Departamento de Biología, Universidad de Los Andes, Venezuela
2Facultad de Farmacia y Bioanálisis, Universidad de los Andes, Venezuela
3Departamento de Química, Universidad de Los Andes, Venezuela
Correspondence: Cristóbal Lárez Velásquez, Laboratorio de Polímeros, Departamento de Química, Facultad de Ciencias, Universidad de Los Andes, Mérida 5101, Venezuela
Received: January 25, 2019 | Published: March 25, 2019
Citation: Rojas M, Peña M, Peña-Vera MJ, et al. Characterization and determination of antimicrobial and metal resistant profiles of Xanthomonas strains isolated from natural environments. J Anal Pharm Res. 2019;8(2):55?60. DOI: 10.15406/japlr.2019.08.00312
Three Xanthomonas strains were isolated from natural environments in Venezuela and studied morphologically, subjected to biochemical characterization and microbiologically and molecularly identified. In order to reconfirm the identity of these strains, the production of xanthan was induced for Xanthomonas campestris and Xanthomonas vesicatoria isolates, obtaining a yield of 7 and 35mg/100mL of culture, respectively; the characterization by FTIR spectroscopy of the obtained products confirmed the formation of xanthan during the growth of the strains. Characterized strains were submitted to resistance studies to 14 commercial antibiotics of clinical use as well as to 10 metal salts were carried out in Luria Bertani (LB) medium, which disturbingly confirmed these Xantho-monas strains present important percentages of resistance to the antibiotics (79%) and to the salts (50%) tested.
Keywords: Xanthomonas, xanthan, resistance profiles, metal resistance, strain identification
Antimicrobial resistance happens when microorganisms such as bacteria, fungi, viruses, and parasites, undergo genetic changes during their exposure to antimicrobial agents, i.e., antibiotics, antifungals, antivirals, antimalarials, anthelmintics, etc., that make them stronger. Microorganisms that develop antimicrobial resistance are currently referred to as “superbugs”. Patients with infections caused by these microorganisms are at increased risk of worse clinical outcomes and death, and additionally consume more health-care resources than patients infected with non-resistant strains of the same bacteria.
On the other hand, the use of antibiotics in agriculture and livestock has been routinely described as a major contributor to the clinical problem of resistant disease in human medicine.1 Xanthomonas is a genus of facultative phytopathogenic bacteria, which developed mainly as parasitic organisms in host plants, and partly in the soil as saprophytes, so that environmental changes can directly affect their survival and cause permanent genetic changes. Studies on Xanthomonas spp. gained importance in agriculture from the moment these microorganisms were identified as the vesicatorium bacteria infecting tomato (1921)2,3 and pepper (1923).4
The genus Xanthomonas is widely known for its phytopathogenic character and its activity on many plants has been widely documented. Conversely, its pathogenicity in humans has been scarcely considered to the point that Xanthomonas campestris has been included in the 2016 updated European Food Safety Authorit (EFSA) list of qualified presumption of safety (QPS) of recommended biological agents for xanthan gum production.5 Nevertheless, it is important to mention that there is a reported clinical case where it microorganism was isolated on 2 separate occasions from the blood of a Chinese 58-year-old woman who had undergone a ventriculostomy for ventricle bleeding. The patient's fever was abated after treatment with erythromycin and chloramphenico.6
Contrary to statements from the FESA report,5 there are numerous papers related to acquisition of antimicrobial resistance by X. campestris7–10 including the pioneer work to identify the bacteria infecting tomato reporting that it is extraordinarily resistant to copper sulphate.2 Due to the historical controversy about of the bacterial resistance acquisition mechanisms,11 this work was carried out to biochemically characterize local Xanthomonas campestris strains, isolated from natural environments and character-ized, and then ascertain their resistances of to some commonly used commercial antibiotics as well as to some metallic salts.
Bacterial strains
Origin of the three bacterial strains studied is shown in the Table 1. These isolates were obtained from different plant species and were identified as genus Xanthomanas only through observation studies with the optical microscope. A deeper characterization was necessary for a complete identification of these Xanthomonas species. This is what gave origin to this work.
Name |
Code |
Origin laboratory |
Host plant |
X. campestris |
2044 |
Micology Laboratory (IVIC)* |
Brassica oleraceavar. Capitata(cabbage) |
X. campestris |
2059 |
Micology Laboratory (IVIC)* |
Phaseolus vulgarisL.(bean) |
X. vesicatoria |
2093 |
Laboratory of Phytopatogenic Bacteria (UCLA)** |
Capsicum annuum(peppers) |
Table 1 Origin of the bacterial strains used in this work
*Instituto Venezolano de Investigaciones Científicas
**Universidad Centro Occidental Lizandro Alvarado
Morphological studies of bacterial strains
Micromorphological study: Consisted in evaluation of cell motility and shape by observation under the optical microscope and determination of the biochemical nature of the bacterial wall (Gram staining).12
Macromorphological study: The macroscopic characteristics of the colonies such as color, shape, texture and appearance, as well as the production of pigments, were observed after 24 hours of incubation of isolates cultured on brain heart infusion (BHI) agar at 30°C.13
Biochemical characterization
A preliminary taxonomic location at the group or family level was performed using the following biochemical tests: Hugh and Leifson medium, nitrate reduction, MIO (Motility, Indol, Ornithine), oxidase test, MacConkey Agar, and urea hydrolysis, according to procedures previously described.12-14
Microbiological identification of Xanthomonas strains
The microbiological identification was performed by analytical profile index (API) tests using an isolated colony of each microorganism. A suspension was prepared in 5mL of sterile water then adding it by filling the tubes, not the dome of all wells (each well had a tube and a dome aerobic part). The dome of the CIT, VP, GEL wells were filled with the bacteria suspension and the domes of the ADH, LDC, ODC, URE, H2S wells were covered with paraffin to obtain anaerobiosis. The strip was placed in its own humid incubation chamber; water was previously added in the wells of the chamber to provide a moist atmosphere during incubation. Incubation was carried out at 37°C for 18-24 hours and, after incubation, immediately recording results that do not need to be revealed. The reading of the results was carried out comparing the colors of each well with those of the reading tables, registering the result as positive or negative. A numerical profile of 7 numbers was obtained from the set of reactions and results.15
Molecular identification of Xanthomonas strain
The strain 2044 was inoculated in Luria Bertani medium (LB) to have fresh strains for the PCR tests. Molecular identification consisted of amplification of the 16S rDNA gene by PCR using the universal bacterial primers 27F (5'-AGAGTTTGATCCTGGCTCAG-3`)16 and 1492R (5'-GGTTACCT TGTTACGACTT-3').17 Amplification reactions were prepared for a total volume of 10μL, containing: 5μL of GoTaq® Green Master mix (Promega), 2μL (0.1μg/μL) of each primer and 4.6μL of sterile distilled water. PCR amplifications were performed on an Applied Biosystems 2720 Thermocycler with the following program: bacterial lysis and denaturation at 95°C for 10 min, followed by 30 cycles of 94°C for 45s, 51°C for 45 s and 72°C for 1.5min per cycle, with a final extension step for 10 min at 72°C.17-19 Subsequently, the salt purification of the amplification products was carried out according to the protocol described by Sambrook and Russell,20 with the following modifications: to the remaining PCR product, after using 3μL for visualization in the gel of agarose, sterile distilled water was added until a volume of 100μL, then a volume of 10 M ammonium acetate and 2.5 volumes of absolute ethanol (100 % v/v) were added. This mixture was allowed to precipitate for 10 min and the supernatant was discarded. The DNA pellet was twice washed with 1mL of 70 % (v/v) ethanol, centrifuging for 10 min and 5 min at 13.4 rcf, respectively. The pellet was then dried at room temperature and resuspended in 11μL of sterile distilled water. In order to verify the presence and quality of the DNA after purification, the precipitated product was resuspended on a horizontal 0.8% agarose gel and visualized with a solution of 50ng/mL of ethidium bromide, loading 1μl of the purified product after combining it with 1μL of 6X loading buffer and 4μL of sterile distilled water.
The purified products were sent to Macrogen Inc. (Korea) for sequencing using the forward primer (27F). Subsequently, contigs were generated using the obtained sequences and the BioEdit Sequence Alignment Editor program.20 Then a similarity analysis between the sequences was performed by comparing the contigs obtained from the Basic Local Alignment Search Tool (BLAST)21 and sequences published in the GenBank databases.22 The bacterial identity was assigned taking into account the results from this comparison, using as identity parameter a "max ident" ≥85 % and a coverage in sequence alignment (Query coverage) ≥98 % on BLAST (Nucleotide Blast) with Nucleotide collection within GenBank (NCBI).23
Antibiotic resistance
Resistance to antibiotics was studied in LB medium prepared with antibiotics commercially purchased on drugstore. Antibiotics, selected in tablet presentations, were Clarithromycin (Cla) 500mg, Levofloxacin (Lvfc) 500mg, Amoxicillin (Am) 500mg, Cephalothin (Cf) 1g, Ceftazidime (Caz) 1g, Gentamicin (Gm) 80mg, Amoxicillin/Clavulanic (Amc) acid 500mg, Cefadroxil (Cfr) 500mg, Ciprofloxacin (Cxm) 500mg, Ampicillin (Amp) 500mg and Kanamycin (Kan) 500mg, Chloramphenicol (Cam) 500mg while those from intravenous presentation were Ceftriaxone (Cax) 1g and Cefotaxime (Cft) 1g. Each of them was diluted to a stock concentration of 100mg/mL.24 From these stock solutions, mLD medium plates (25mL) were prepared with the following concentrations of each antibiotic: 25, 50, 100 and 200μg/mL. The strains were then inoculated using chopsticks and incubated at 37°C, taking readings at 24 and 48h of incubation. Each experiment was performed in triplicate.
Metal resistance
Growth of Xanthomons strains in LB plates, separately supplemented with concentrations of 2 and 5mM of the different metals, was studied following the methodology used in other studies,25 with some modifications. Metallic cations (salts) tested were: Ni2+ (NiSO4), Hg2+ (HgCl2), Cu2+ (CuSO4), Sn2+ (SnCl2), Ag2+ (AgNO3), Fe3+ (FeCl2), Ba2+ (BaCl2), Zn2+ (Zn(CH3CO2)2), Li (LiOH) and Mn2+ (MnCo3). The procedure in order to observe the strain growth in Petri dish can be briefly described as follow: 100µl of the bacterial suspension (previously prepared in liquid LB medium) were inoculated and incubated in the supplemented media at 37°C, taking the reading at 24 and 48h of incubation. Each experiment was performed in triplicate.
In order to establish the Minimal Inhibitory Concentration (MIC) for each of the metals used in the previous analyzes, strains of Xanthomonas were analyzed with an LB broth supplemented with different concentrations of each of the metals using sterile test tubes.
Each tube, containing 5mL of broth supplemented with a metal at a given concentration (ranging between 1, 2, 4 and 4mM), was inoculated with a 140μL aliquot of the correspondent strain in the late exponential phase, at an initial optical density at 600nm (OD600) of 0.03, and the plates were incubated at 37°C for 24- 48h. Each experiment was performed in triplicate.
Induction of xanthan gum production
Bacterial strains were inoculated on medium 1 (sucrose 20g/L, peptone 5g/L, K2HPO 0.5g/L, MgSO47H2O 0.25g/L, agar 16.0g/L) plates, and allowed to grow between 18-20 hours. After growth, a colony was taken from medium 1 to a cassette with 10mL of yeast medium (YM) broth or medium 2 (yeast extract 3.0g/L, malt extract 3.0g/L, peptone 5.0g/L, glucose 20g/L), allowing to grow from 5 to 7 hours. Subsequently, 5 to 10% of the volume of a culture of 100mL of induction medium or medium 3 (NH4H2PO4 1.5g/L, K2HPO4 2.5g/L, MgSO47H2O 0.2g/L, sucrose 40g/L) was transferred into a 250mL cassette. It was allowed to grow at 28°C in shaking at 180rpm for 48hours. Subsequently, the culture was centrifuged at 12,000rpm for 30 minutes at 4°C. Ethanol was added to the supernatant in a 1:4 (v/v) ratio and left overnight at 4°C. The next day the mixture was centrifuged at 10,000rpm for 30 minutes at 4°C to recover the precipitate, which was washed with 100mL of water and heated at 60°C for 30 minutes. Thereafter, ethanol was added in the same ratio 1:4 and centrifuged again. Finally the precipitate was recovered and allowed to dry in the oven at 40-45°C overnight.26
Xanthan FTIR spectroscopy study
The FTIR from precipitate obtained was performed in a Perkin-Elmer RX1 IR-TF spectrometer. The solid sample was mixed with pure KBr in a mortar until to obtain a fine powder. A small portion of this powder was placed in a sample carrier to prepare discs using a hydraulic press.
Strain characterization
By direct observation under a microscope, the three strains studied in this work were found to be bacillus with positive motility and Gram negative staining. Macro-morphological analysis of them, in BHI medium, allowed the determining of colonies of abundant extension, punctate shape of 1 mm in size, with full borders, opaque density, viscous consistency and light yellow color.
Strains were subjected to a battery of biochemical tests (oxidase, KIA, LIA, citrate, MIO, indol-motility and urea) in order to obtain a preliminary taxonomic location and type of API gallery that should be used for taxonomic identification by means of microbiological techniques. All the Xanthomonas isolates were negative for all the biochemical assays tested. The API 20E gallery was chosen for Gram-negative bacilli outside the family Enterobacteraceae, which allow genus and species identification. According to results obtained, the 2044 and 2059 strains they were identified as Xanthomonas campestris, with a high percentage of reliability of 90.4 y 92.7 %, respectively. In the case of strain 2093, it was identified as Xanthomonas vesicatoria, with 91.5% reliability (Table 2).
Strain |
API code |
Bacterial identification |
Reliability percentage (%) |
Bergueyʼs manual |
2044 |
2671205 |
X. campestris |
90.40 |
24/26 |
2059 |
0472341 |
X. campestris |
92.70 |
23/26 |
2093 |
0473300 |
X. vesicatory |
91.50 |
24/26 |
Table 2 API 20E tests performed on the different Xanthomonas strain
A molecular identification by PCR amplification of the Xanthomonas strains was performed, using the 27F and 1492R primers. Figure 1 shows the 1500bp amplification corresponding to the gene 16S DNAr of strains 2044, 2059 y 2093. A higher quality after purification can also be assumed due to the observation of a simple band in this electrophoretic profile.
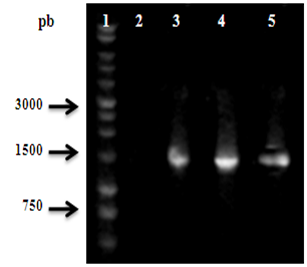
Figure 1 PCR amplification of the 16S DNAr gene of the isolated Xanthomonas strains. The position at ~1500bp corresponds to 16S DNAr gene. 1: Molecular weight marker; 2: Negative Control; 3-5: Isolates 2044, 2059 and 2093, respectively.
The 16S DNAr sequences were compared with sequences from the GeneBank databases employing the BLASTn bioinformatic program (Table 3). This table shows the percentage of identity with respect to the phylogenetically closest species for each of the strains studied. It can be seen that the isolates 2044 and 2059 were identified as Xanthomonas campestris. In reference to isolate M2093 it was identified with Xanthomonas vesicatoria. Identity percentages between 95 and 96% were obtained. These results coincide with the results obtained in the microbiological tests.
Strain |
Species phylogenetically closer |
Nº Access |
% Identity |
2044 |
X. campestris strain IR-Isfahan (B) |
KF964554 |
95 |
2059 |
X. campestris pv. Campestris strain 8004 |
NC_007086.1 |
96 |
2093 |
X. vesicatoria strain ATCC 35937 |
NR_026388.1 |
96 |
Table 3 Comparative analysis of the nucleotide sequence of the 16S DNAr gene of the different strains of Xanthomonas isolated
In order to reconfirm the identity of the studied strains, the production of the biopolymer xanthan was induced by growth of the 2044 y 2093 strains in the induction medium. From the culture of each of these strains, a yellowish powder was obtained with a yield of 7 and 35mg/100mL of culture, respectively. Obtained xanthan samples were chemically characterized by means of FTIR spectroscopy with the purpose of confirm the presence of the characteristic xanthan chemical groups. Figure 2 shows the xanthan chemical structure and two typical spectra obtained for samples obtained from of 2044 and 2093 strains, which perfectly agree with previously reported spectra for xanthan obtained from X. campestri strains.27-29 The more prominent bands can be associate to: O-H stretching from hydroxyl groups (3410cm-1); C-H stretching from aliphatic CH2 and CH3 moieties (2935cm-1); C=O stretching from carboxylic and ester moieties (1670cm-1); C-O-C st retching from ether linkages (1060cm-1) and skeleton and complex ring vibrations (570cm-1).
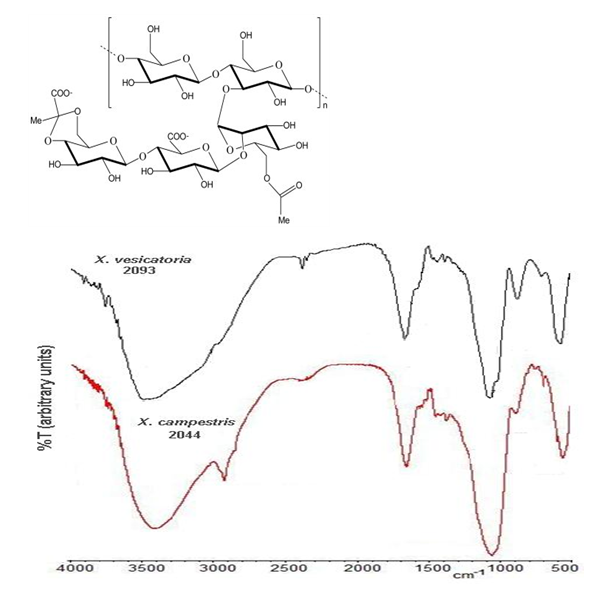
Figure 2 Structure and FTIR spectrum of the xanthan samples obtained during growth of the 2044 and 2093 strains in medium 1; FTIR samples were prepared as KBr discs.
Antibiotic and metal resistant studies
The study of antibiotic resistance was carried out using commercial antibiotics prepared at different concentrations. Results demonstrate that all Xanthomonas isolates have resistance to different antibiotics. Thus, X. campestris 2044 and 2059 strain have resistance to 11 y 10 antibiotics, respectively, yet at a concentration as low as 50μg/mL (Table 4). It can also be observed that these strains exhibited resistance for Am, Can, Amp, Cf, Kam and Cax at all concentrations tested while for Amc, Gem, Caz, Cft and Cfr were necessary concentrations higher than 50μg/mL in order to inhibit the bacterial growth. On the other hand, strain 2044 y 2059 was found to be sensitive to the antibiotics Cxm, Lvfc and Cla. In general, a resistant behavior of this two isolates is observed to more than 10 antibiotics studied.
Antibiotics (µg/mL) |
Isolated |
Antibiotics (µg/mL) |
Isolated |
||||||
|
X.c 2044 |
X.c 2059 |
X.v2093 |
|
X.c 2044 |
X.c 2059 |
X.v2093 |
||
Amp |
25 |
R |
R |
S |
Amc |
25 |
R |
R |
S |
50 |
R |
R |
S |
50 |
R |
R |
S |
||
100 |
R |
R |
S |
100 |
R |
R |
S |
||
200 |
R |
R |
S |
200 |
S |
S |
S |
||
Cfl |
25 |
R |
R |
S |
Am |
25 |
R |
R |
S |
50 |
R |
R |
S |
50 |
R |
R |
S |
||
100 |
R |
R |
S |
100 |
R |
R |
S |
||
200 |
R |
R |
S |
200 |
R |
R |
S |
||
Kan |
25 |
R |
R |
R |
Gm |
25 |
R |
R |
S |
50 |
R |
R |
R |
50 |
R |
R |
S |
||
100 |
R |
R |
R |
100 |
R |
R |
S |
||
200 |
R |
R |
R |
200 |
S |
S |
S |
||
Cax |
25 |
R |
R |
S |
Caz |
25 |
R |
S |
S |
50 |
R |
R |
S |
50 |
R |
S |
S |
||
100 |
R |
R |
S |
100 |
S |
S |
S |
||
200 |
R |
S |
S |
200 |
S |
S |
S |
||
Lvfc |
25 |
S |
S |
S |
Cxm |
25 |
S |
S |
S |
50 |
S |
S |
S |
50 |
S |
S |
S |
||
100 |
S |
S |
S |
100 |
S |
S |
S |
||
200 |
S |
S |
S |
200 |
S |
S |
S |
||
Cla |
25 |
S |
S |
S |
Cft |
25 |
R |
R |
R |
50 |
S |
S |
S |
50 |
R |
S |
S |
||
100 |
S |
S |
S |
100 |
S |
S |
S |
||
200 |
S |
S |
S |
200 |
S |
S |
S |
||
Cfr |
25 |
R |
R |
S |
Cam |
25 |
R |
R |
R |
50 |
R |
R |
S |
50 |
R |
R |
S |
||
100 |
R |
R |
S |
100 |
R |
S |
S |
||
200 |
S |
S |
S |
200 |
R |
S |
S |
||
Table 4 Results of the resistance study of X. campestris M2044 strain to antibiotics salt
R, resistant; S, susceptible
In the other hand, the Xanthomonas vesicatoria 2059 strain showed greater sensitivity to antibiotics. It was resistant only to 3 antibiotics. This marked difference between the antibiotic resistance profiles of the X. campestris isolates (2044/2059) and X. vesicatoria (2093) could be attributed to the difference in the amount of xanthan synthesized by these isolates. In the case of X. vesicatoria, xanthan could be acting as a barrier to the acquisition of some antibiotic resistance genes through horizontal gene transfer (HGT). It is known that the presence of extracellular polymers in the bacterial surface can act as a barrier to some HGT processes.30,31
As can be seen from the Table 4, locally isolated strains of X. campestris showed resistant in a high percentage (~79%) to the antibiotics tested (14). Because the tested antibiotics are of extended clinical use, the presence of these strains could represent a risk in such areas due to the potential transmission of their resistance to other bacteria as it has been reported to occur in other environments.32
Similarly, in order to evaluate the resistance of 2044, 2059 and 2093 Xanthomonas strains to different metals, these were cultivated in LB medium supplemented with different salts of metals, studying their growth capacity in the different quadrants of the Petri dish. In the results shown in Table 5, it can be appreciated that Xanthomonas isolates showed resistance to five metals until a concentration of 5mM (Table 5). The MIC values of each metal were evaluated in liquid medium for each of the Xanthomonas isolates (Table 6) considering that resistance values obtained by the culture of isolates on agar media supplemented with metals can cause an overestimation of this parameter. When considering the combined results of both metal resistance studies (plaque and liquid medium), it is clear that three Xanthomonas isolates could grow in the presence of 4mM of at least five of the ten metals tested.
|
Metals (mM) |
Isolated |
|||
|
X.c 2044 |
X.c 2059 |
X.v 2093 |
||
|
Ba2+ |
2 |
R |
R |
R |
|
5 |
R |
R |
R |
|
|
Li+ |
2 |
R |
R |
R |
|
5 |
R |
R |
R |
|
|
Zn2+ |
2 |
R |
R |
R |
|
5 |
R |
R |
R |
|
|
Cu2+ |
2 |
R |
R |
S |
|
5 |
S |
S |
S |
|
|
Ni2+ |
2 |
R |
R |
R |
|
5 |
R |
R |
R |
|
Table 5 Heavy metal resistance for the different isolates of Xanthomonas tested in agar medium
R, resistant; S, susceptible
|
Metals (mM) |
Isolated |
|||
|
X.c 2044 |
X.c 2059 |
X.v 2093 |
||
|
Ba2+ |
1 |
R |
R |
R |
|
2 |
R |
R |
R |
|
|
3 |
R |
R |
R |
|
|
4 |
R |
R |
R |
|
|
Li |
1 |
R |
R |
R |
|
2 |
R |
R |
R |
|
|
3 |
R |
R |
R |
|
|
4 |
R |
R |
R |
|
|
Zn2+ |
1 |
R |
R |
R |
|
2 |
R |
R |
R |
|
|
3 |
R |
R |
R |
|
|
4 |
R |
R |
R |
|
|
Cu2+ |
1 |
R |
R |
R |
|
2 |
R |
R |
R |
|
|
3 |
R |
R |
R |
|
|
4 |
R |
R |
R |
|
|
Ni2+ |
1 |
R |
R |
R |
|
2 |
R |
R |
R |
|
|
3 |
R |
R |
R |
|
|
4 |
R |
R |
R |
|
Table 6 Heavy metal resistance among the different isolates of Xanthomonas tested in liquid medium
On the other hand, sensitivity was found at all the concentrations tested in five of the metallic salts studied (mercuric chloride, tin chloride, silver nitrate, manganese carbonate and iron chloride). Moreover, the X. campestris strain 2044 is disturbingly resistant to metals associated with human toxicity and representing the major components of pesticides, such as lithium, barium and copper.
It was possible to characterize local strains of Xanthomonas through micro- and macro-biological tests, establishing their taxonomic location through biochemical and microbiological tests. For the three Xanthomonas isolates (2044, 2059, 2093) the coding sequence of the 16S DNAr gene was amplified by PCR. The isolates studied in this work belong to the species X. campestris and X. vesicatoria. To reconfirm the identity of these strains, the production of xanthan was induced, obtaining a yield of 7 and 35 mg/100mL of culture for X. campestris and X. vesicatoria, repectively. Characterization by FTIR spectroscopy of the obtained xanthan clearly confirmed its formation during the strains growth.
Xanthomonas isolates showed resistant in a high percentage (79%) to the commercial clinical antibiotics tested (14). Similarly, studies with metallic salts showed resistance to 50% of the salts tested (10), which is also a fact worthy of attention because many of the pesticides used in agriculture are based on this type of compound.
Results obtained in this study clearly demonstrate that Xanthomonas evaluated in this work, which were isolates from natural environments, present multi-drug resistance.
None.
Authors declare that there are no conflicts of interest.

©2019 Rojas, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.