Journal of
eISSN: 2473-0831


Research Article Volume 3 Issue 4
Correspondence: Nagarajan Kayalvizhi, Department of Zoology, School of Life Sciences, Periyar University, Salem, Tamil Nadu, India
Received: October 21, 2016 | Published: November 4, 2016
Citation: Selvam D, Thangarasu A, Neelamegam R, Muthukalingan K, Nagarajan K (2016) Antimicrobial Potential of Earthworm Wegeneriona sp. against Human Pathogens. J Anal Pharm Res 3(4): 00060. DOI: 10.15406/japlr.2016.03.00060
The medicinal value of earthworms has been known for centuries. The earthworms are the source of proteins, peptides, enzymes and physiologically active substances. Thus in the present study, the earthworm skin extracts were prepared and subjected to antimicrobial activity which was determined by the agar well diffusion method. The antimicrobial substances showed activity against Serratia marcescens. The molecular weight of the peptide from skin extract was found to be ~3kDa by SDS – PAGE and confirmed by zymogram analysis. Further the earthworm was identified by morphological and COI gene sequence analysis and resulted to be Wegeneriona sp. This study may lead to the formulation of new antimicrobial drug from the earthworm skin extract.
Keywords: antimicrobial peptide, coi, serratia marcescens, wegeneriona sp, sds-page, zymogram, antimicrobial
The earthworm is distinctive from saprozoic organisms. Earthworms are ancient creatures, which have survived on apex of the earth at least 700 million years ago.1 Earthworms are usually precise the same as a relatively well known group of soil invertebrate from both ecological and taxonomical perception. Earthworms are enforced to adopted and survive in this type of environments.2 The earthworm skin play a key role in the everyday survival of amphibians and their capability to develop a wide range of habitats and ecological conditions, and act as the first line to against harmful aggression by microorganisms, parasites, and predators. Hence, they are expected contains to excellent chemical defense system composed of pharmacological and antimicrobial peptides. Several anti-infection components including lysozymes, fetidins, and antimicrobial peptides have been originated in earthworms. More than 3000 species earthworm are known, only three antimicrobial peptides are identified from three species.1,3,4
Antimicrobial peptides (AMPs) are fashioned by multicellular organisms. They are defense mechanism for against competing pathogenic microorganism.5,6 Antimicrobial peptides (AMPs) represent a wide range of category to identified in different families of extremely conserved peptides widely found throughout nature, which demonstrate broad-spectrum antimicrobial activity in vitro and in vivo.7 Many peptides are identified by purification of the peptides through antibiotic activity, even though through other species, the increased use of bioinformatics techniques allowed to identify more number of peptides.8 The earthworms have medicinal components which are in Chinese medicine. Nowadays, the developments of earthworm medical value contemplate on fibrinolytic enzymes and antibacterial proteins. Agglutinins have been found in skin secretions, coelomic fluid and earthworm tissues.9 In the present investigation has been focused on isolation and identification of earthworm species by COI gene sequencing method. The antibacterial peptide from the earthworm skin showed inhibition against Serratia marcescens and SDS-PAGE confirmed the molecular weight of antimicrobial peptide.
Collection of earthworm
The earthworms were collected from Muthunaickenpatti (Salem, Tamil Nadu) (11°42’24.2”N; 78°01’40.1”E) near paddy field. The worm would be view in surface area to 2 inches depth to collect the earthworm. The worms were washed in running tap water in order to remove the sand particles from the surface of earthworm.
External reproductive morphology
Earthworms were taken in a small beaker and anaesthetized using chloroform, After 5 minutes, the earthworms were taken into chloroform, earthworms were pinned dorsally and observations were made on the external morphological and characters of the mouth and anterior segments, the position of the spermathecal openings; the clitellar segments and the female and male genital openings or apertures. Observations were also made on inter segmental grooves the dorsal pores and the arrangement of setae.
Preparation of earthworm extract
Mature earthworms (average body weight 0.3 to 0.5g) were washed with running tap water to remove any dust from body surface. The earthworms were kept in 0.9 % NaCl at room temperature for 1-2h with few changes of solution until their digestive system was clean. 3g of earthworm tissue were homogenized in 1 ml of phosphate buffer solution (pH 7.2). Homogenized solution was collected in 1 ml centrifuge tube and centrifuged at 10,000 rpm at 4°C for 15min. After the supernatant was collected the new centrifuge tube. Then supernatant was used to determine the antimicrobial activity.
Genomic DNA extraction from earthworm skin
Earthworm tissues up to 1 cm were taken in clean and dry eppendorf tube, then added 200μl of 1xTE buffer (1M Tris Hcl, 0.5 M EDTA bring upto volume 1 L double distilled H2O was suggested by Doyle & Doyle). After that 500 μl CTAB buffer were added (1M Tris Hcl, 5M NaCl, 0.5M EDTA 20% Cetyltrimethyl ammonium bromide (CTAB) to bring total 1L volume of double distilled H2O followed.10 After that 3-5μl of Proteinase K (20mg/ml) added and vigorously vortex. Added RNase (2μl) (10mg/ml) and incubate at -20°C for 1hour or overnight. (RNase was also added to prevent RNA contamination). This sample was incubated at 55°C for 1 hour, mixed intermittently. Added equal volume of Chloroform: Isoamylalcohol (24:1). Centrifuged at 14000rpm for 7min. Transferred aqueous phase to a fresh centrifuge tube and above step repeated twice, added 35μl of chilled 7.5M ammonium acetate, 200μl isopropanol (0.5 volumes), and left DNA to settle down for 20 minute. Centrifuge at 14000 rpm for 10 minute. This supernatant was discarded, added 500μl 70% ethanol. Centrifuge at 14000 rpm for 3minute, discarded the supernatant. Added equal volume of the 100% ethanol. Centrifuge at 14000 rpm for 3minute, discarded the supernatant. The tube was kept for drying at room temperature dry pellet or DNA dissolve in 30μl 1X TE (10 mM Tris HCl and 1 mM EDTA) buffer and incubated for 2 hour at room temperature. Stored at -20°C for further use.
PCR amplification
PCR amplification was carried out in a 1µL of template DNA in 20µL of PCR reaction solution. The initial denaturation for 5min at 94°C and then 35 cycles of denaturation for 45 sec at 94°C, annealing for 60sec at 55°C, and extension for 60sec at 72°C with a final extension at 72°C for 10min. A fragment of the mitochondrial cytochrome c oxidase subunit I sequence (COI) was amplified using the primers HCO2198 (5’ – TAAACTTCAGGGTGACCAAAAAATCA-3’) and LCO1490 (5’ – GGTCAACAAATCATAAAGATA TTGG- 3’) 11. 10 µL PCR products were electrophoresed on 1% agarose gel to determine the size of product. The phylogenetic tree was constructed using CLUSTAL-W program of biology work surface.
Test microorganisms for antimicrobial studies
The microbial strains were obtained from the IMTECH, India. Were tested against gram positive bacteria, Staphylococcus aureus (MTCC 96), Bacillus aryabhattai (MTCC 14579), Bacillus megaterium (MTCC 441), and gram negative bacteria such as Serratia marcescens (MTCC 4822), Klebsiella pneumonia (MTCC 4727), Pseudomonas putida (MTCC 1194) were grown in a nutrient broth at 37°C for 12h.
Antimicrobial activity assay
Antimicrobial activity of earthworm extract was carried out by using agar well diffusion method.12 Luria broth used for bacterial cultures was inoculated with broth, suspended separately. The cultures were swabbing in the agar and the plates were left for solidification. The total of 8mm diameter wells was punched into culture swabbing plate. The wells were filled with earthworm extraction and they are completely dissolved in the sample. The plates were incubated at 37°C for 18h. The antibacterial activity was determined by measuring the diameter of zone of inhibition.
SDS – PAGE and Zymogram analysis
SDS-PAGE was performed according the method of13 electrophoretic apparatus. 12% separating and 4% stacking gel were used to separate the proteins. Gel electrophoresis was carried out with SDS and β-mercaptoethanol in the loading buffer. A sandwich was made with two glass plates separated by Spacer Strip (1.5mm thickness). Loading buffer of pH 6.8 (60mM Tris base, 5% SDS, 1% β- mercaptoethanol, 0.002% bromophenol blue and 6% glycerol) was added to the sample. Per well 50μl samples were loaded. SDS-PAGE was run until the bromophenolblue front reached the end of the gel (30 V for stacking gel and 50 V for separating gel was applied). After completion of electrophoresis gel was stained with coomassie brilliant blue and destained subsequently with excess of methanol in destaining solution. The zymogram analysis for gel was water washed 2 to 3 h, and then nutrient agar plate was prepared. 0.75% soft agar preparing then seeded S. marcescens. The plate was incubated 37°C for 24 h.
Morphology
The body of the clitellar earthworm, its light gray in colour, round or cylindrical in shape and metamerically segmented. The length 80-110 mm; diameter 3-5mm (Figure 1). The arrangement of setae is very strong. The anterior region shows the prostomium which is epilobus. On the ventrolateral surface, the body segments 8, 9, 10, 11 and 12 shows the presence of spermathecal openings. Exactly on the 12 segment, a pair of female genital openings is present. Segments 13, 14 and 15 forms the unsegmented collar like structure called the clitellum. On the 16 segment, pair of male genital openings is present, bellow the clitellum.
The COI sequencing analysis
The genomic DNA was isolated from the earthworm tissue and subjected to amplification using COI specific universal primer (Figure 2). A fragment of 649 bp was amplified and photographed (Figure 3). The sequencing analysis revealed that the earthworm belong to Wegeneriona sp. In order to find the relatedness to other organisms phylogenetic tree was constructed by maximum likelihood analysis and results showed closeness towards Wegeneriona sp (Figure 4). Taxonomy and molecular techniques were used to express in a taxonomically included technique of new species of earthworm. COI sequences for detecting species level differences even in cases of recent divergence. Furthermore, they indicate that COI divergence may help to clarify misleading morphological classifications.14 The DNA sequencing of holotype helps the account of a new species by confirm that the taxonomically diagnostic matches with its position in the phylogeny. In addition, prospective incongruence is easily visible on a phylogenetic tree. This study makes available confirmation that a COI – based identification system would be successful for identifying earthworm species.
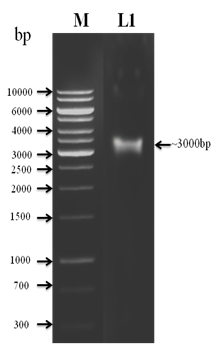
Figure 2 Isolated Genomic DNA from earthworm tissue to run in 1% agarose gel electrophoresis containing Ethidium bromide. DNA photograph was taken under transilluminator emitting UV light.

Figure 3 PCR amplification of Cytochrome c Oxidase (COI) gene using specific primer were used for the amplification.
Antimicrobial activity
The antimicrobial function of earthworm skin extraction in response to Serratia marcescens, Staphylococcus aureus, Klebsiella pneumonia, Bacillus aryabhattai, Bacillus megaterium and Pseudomonas putida were detected using the agar well diffusion assay method. The antimicrobial function of earthworm skin extraction to S. marcescens exposure was highest and the diameter of the growth of inhibition was 3mm (Figure 5); whereas S. aureus, K. pneumonia, B. aryabhattai, B. megaterium, P. putida does not have the activity. Antimicrobial peptides, which are now considered as one of universal host defense tools of living organisms against microbial infection,15 have been found in both vertebrates and invertebrates. Most of antimicrobial peptides so far found in invertebrate were from insects,16 crustaceans17 and nematodes.18 We describe here the antimicrobial peptide from the earthworm, which showed antimicrobial activity against a broad range of microorganisms. This is one of the reports on the annelid antimicrobial peptide which does not possess any hemolytic activity. The extraction showed maximum activity in response to S. marcescens.
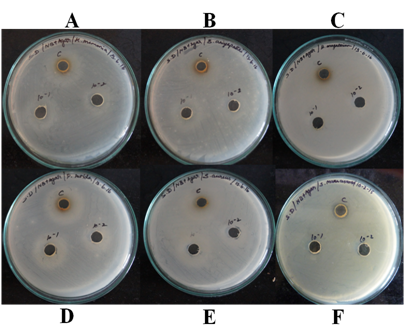
Figure 5 Minimum inhibitory concentrations for earthworm skin extraction AMP against A. Klebsiella pneumonia, B. Bacillus aryabhattai, C. Bacillus megaterium, D. Pseudomonas putida, E. Staphylococcus aureus, F. Serratia marcescens, with different concentration. S. marcescens is only clear zone for appear.
SDS-PAGE and zymogram analysis
A coomassie briliant blue stained SDS-PAGE gel revealed a diffuse band of the concentrated crude sample (Figure 6). A comparison of the band on the stained gel with the half gel showing inhibition zones indicates that band contained the antimicrobial activity. This zone of inhibition was observed in ~3kDa, which represents the antimicrobial peptide.
In particular, this approach allows species identification for specimen fragments that often form a significant proportion of ecological samples and cannot be reliably identified with morphological characters. The current development come out to be that DNA barcoding requirements to be used at the side of traditional taxonomic tools and different forms of molecular systemic so that problem in cases can be identified and errors detected. This study clearly indicates that the earthworm tissue extract to formulation of new natural antimicrobial agent and thus may found beneficial in future prospects for mankind.
The authors gratefully acknowledge the Department of Science and Technology, New Delhi for providing partial financial support under DST-WOS-A (DST/SR/WOS-A/LS-629/2012(G) Scheme.
The authors declare no conflict of interest.
None.

©2016 Selvam, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.