Journal of
eISSN: 2473-0831


Human tyrosinase (Tyr) is a Type I membrane glycoprotein that is the rate-limiting enzyme for controlling the production of melanin pigment in melanosomes. Currently, ~300 Tyr mutations are known to be involved in the genetic disease oculocutaneous albinism type 1 (OCA1), which exists in two forms, OCA1A and OCA1B. OCA1A is caused by a full loss of Tyr enzymatic activity, resulting in the absence of pigment in the skin, hair, and eyes, while OCA1B has reduced Tyr activity and pigment. Here, we used molecular modeling to try to understand the role of genetic changes at the protein level in inherited disease. The significant part of Tyr intra-melanosomal domain and five OCA1 mutant variants were built by homology modeling, glycosylated in silico, and refined using molecular dynamics in water. The modeling confirmed experimental results that N347 and N371 glycosylation is vital for protein stability. The changes caused by the T373K mutation indicate a significant impact on protein structure, as expected for OCA1A. In addition, evaluation of free energy changes in OCA1B mutants showed a strong association with the changes observed in our unfolding/refolding experiments in vitro. In conclusion, our results could be useful for understanding the role of OCA1 mutant variants in melanin pigment production, in silico searching for inhibitors and activators of tyrosinase activity, and genotype-to-phenotype analysis in OCA1.
Keywords: human tyrosinase, homology modeling, albinism-causing mutations
TYR, tyrosinase; ER, endoplasmic reticulum; VMD, visual molecular dynamics; RMSD, root-mean square deviation; PSF, protein structure file; 3-HAA, 3-hydroxyantranilic acid
Human tyrosinase (Tyr) is a copper-containing glycoenzyme that catalyzes the rate-limiting first step in melanin production. Its gene (TYR) is mutated in many cases of OCA1, an autosomal recessive disease that can lead to childhood blindness. OCA1 is further categorized by phenotype into type 1A and 1B. OCA1A is characterized as complete loss of Tyr function, while OCA1B exhibits reduced Tyr enzymatic activity. Visible signs of OCA1 include a reduction or absence of pigment in skin, hair, and eyes. The long-term effects of the mutation include sensitivity to UV radiation, predisposition to skin cancer, and visual problems like nystagmus, strabismus, and photophobia. The intra-melanosomal domain of human tyrosinase (TyrD, residues 19–469) and two OCA1B related temperature-sensitive mutants, R422Q and R422W were successfully expressed in insect cells and produced in T. ni larvae.1 In this work the short trans-membrane fragment and tyrosinase cytoplasmic tail were deleted to avoid potential protein insolubility, while preserving all other functional features of the enzymes. Recently, the TyrD and OCA1-related mutant variants, R422W, R402Q, R422Q, P406L,T373K, and R77Q, were expressed in larval biomass, purified and characterized to suggest a direct link between protein stability and loss of pigmentation in OCA1B albinism.2 Analysis of the consequences of tyrosinase deglycosylation demonstrated the reduced protein expression and enzymatic activity due to possible loss of protein stability and protein degradation.3 Although, the full-length human tyrosinase was recently purified and biochemically characterized,4 the atomic structure of Tyr is unknown. Recently the crystal structure of truncated human tyrosinase-related protein 1 was determined at 2.3Å resolution.5 However, attempts at crystallography of truncated human tyrosinase have been unsuccessful.6 While crystal structures are available for many bacterial and fungal tyrosinase species, an accurate human tyrosinase structure is needed to answer current questions related to mutations associated with OCA1.
Much less is known about the role of Tyr glycosylation. Tyr is an N-linked glycoprotein which relies on glycosylation for its proper folding.7,8 While not all glycosylation sites on Tyr have the same effects on folding and activity, the glycosylation site at N371 is essential for normal enzymatic activity.2 Proper folding of Tyr and the ability of its active site to be able to absorb copper also depends on Tyr’s early interaction with two chaperones, calreticulin, and calnexin.7,8 Tyr is a Type I single-pass membrane protein that is synthesized in the endoplasmic reticulum (ER), where it acquires the correct folding conformation before moving to the Golgi apparatus and eventually, the melanosome.7,8
In OCA1A, Tyr is degraded in the ER.7,8 Experimental studies on the T373K mutant showed that loss of the glycan at N371 prevents Tyr from leaving the ER, thus creating an OCA1A disease phenotype.7 In OCA1B, the stable fraction of mutant Tyr with the decreased enzymatic activity is able to move along its normal melanin biosynthetic pathway in melanocytes, suggesting a partial change in protein stability or protein folding.9 In addition, some OCA1B mutations have been shown to be temperature-sensitive. These mutants are less active catalytically and the differences are associated with conformational perturbations in secondary structure of tyrosinase possibly because of partial (localized) protein unfolding.1 The overall phenotype of OCA1 is white hair and white skin at birth. However, an initial diagnosis of OCA1A may later change, depending on DNA analysis, and the development of some pigment later in life remains possible.10,11 Missense mutations are the most common types of mutations associated with OCA1. Other mutation types do occur but are more difficult to analyze when combined with an allele that produces some Tyr activity. Gene sequencing of the TYR gene often reveals an inherited mutation on two alleles: one from the maternal side and one from the paternal side.10,12,13 However, despite OCA1 being an autosomal recessive disease, some sequencing results reveal only one mutated TYR allele. Much research continues to explore the possibility of currently unknown non-TYR genes that may contribute to Tyr deficiency, including the TYR-like pseudo gene.11 Based on the protein sequence of Tyr, there are predicted to be several functional domains, including an epidermal growth factor (EGF)-like domain, a catalytic domain, and a trans-membrane (TM) domain at the C-terminus.1,5 The functional role of the EGF-like domain is unknown. Monophenolase and diphenol oxidase activities are associated with the catalytic domain, which contains two copper ions coordinated by 6 histidine residues at the catalytic site.14 However, the crystal structure of human tyrosinase, which could establish a link between protein structure and function, is currently unavailable.
With a goal to address the relationship between Tyr’s structure and function, we used homology modeling and molecular dynamics computer algorithms to further understand Tyr’s function and computationally verify our experimental data.1–4 We built a model of the significant part of intra-melanosomal domain of human tyrosinase (TyrD), approximately mimicking the structure of an experimental domain engineered as described by Dolinska et al.1 This model was used to analyze the OCA1 disease-causing mutations, T373K, P406L, R402Q, R422Q, and R422W. For this purpose, computational free energy changes between the wild-type and OCA1B mutants were calculated and these results were compared with experimental free energy changes from unfolding/refolding experiments,2 demonstrating a correlation between the experimental and computationally predicted values. This correlation will allow us to computationally predict the severity of known OCA1 mutations.
Molecular modeling
A homology model of TyrD was built similar to that of described earlier,1 refined, and glycosylated in-silico. TyrD’s structure and trajectory files were analyzed using Visual Molecular Dynamics (VMD).15 The root-mean square deviation (RMSD) of the protein backbone was calculated using the NAMD Trajectory Tool in VMD (Figure 1A). Various frames were analyzed for overall errors, improvements, and energies and then selected for evaluation of mutant free energy changes (ΔΔG).
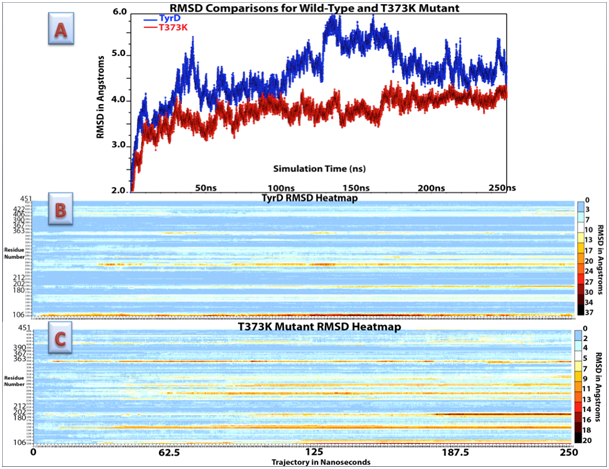
Figure 1 RMSD changes for TyrD and T373K from 0ns to 250ns of molecular dynamics runs. A) Graph of RMSD values over the total trajectory time. B) Heatmap plot of RMSD for individual amino acid residues in TyrD. The color key ranges from the smallest movements in blue to the largest movements in dark red. Highly conserved regions are marked on y-axis key. C) Heatmap plot of RMSD for individual amino acid residues in the T373K mutant. Highly conserved regions are marked on y-axis key. RMSD values are shown in Å.
Mutations were selected based on simultaneous experimental studies evaluating ΔΔG for protein unfolding and secondary structure improvement. An optimal frame of TyrD was chosen, shown to have a high correlation between experimental and computational ΔΔG, and was used to evaluate changes for albinism-causing mutations. The total energy at each nanosecond was extracted from the NAMD output log file and graphed. RMSD values were then compared with total energy fluctuation over each nanosecond of simulation time. Furthermore, a heatmap was created to document protein regions with higher RMSD (Figure 1B). Optimal pdb frames for TyrD represented by 54ns and 122ns frames were chosen for further evaluation because both were in a stable or decreasing RMSD region and at a lower point of total energy, equal to -205,600kcal/mol.
Protein glycosylation
The FASTA sequence for human tyrosinase was downloaded from the UniProtKB database (# P14679)16 and truncated to remove a trans-membrane helix in a similar way that described previously by Dolinska et al.1 Next, the SWISS-MODEL algorithm17 was used to compare the sequence alignment and structure with the top ten most closely matched protein structures found in the Protein DataBank (PDB).18 The following crystallographic structures of tyrosinases from different species were obtained from the PDB: 3NM8, 2AHK, 2AHL, 4J6T, and 3AWZ. A template for the Tyr active site19 was obtained from the Protein Model Database (PM0079416).20 Unfortunately, tyrosinase related protein 15 was excluded from the modeling, because it show no similarity in enzymatic activity with Tyr and the predicted model show the active site instability after 10ns equilibration in water. Finally, a 3D model of TyrD was built for residues 106-451 using homology modeling in the program Yasara (http://www.yasara.org/). Using the program UCSF Chimera, an Extensible Molecular Modeling System,21 a hydroxide ion was added between the two copper atoms to more accurately match the active site model (PM0079416) and to reflect experimental studies depicting the likelihood of a hydroxide ion present in Tyr’s met form.
Next, TyrD was glycosylated at 5 N-glycosylation sites using the online web tool Glycam-Web.22 The exact sugar composition in each N-glycosylation site of TyrD is not known.1 Therefore, the most biologically common eukaryotic core glycan was chosen: (N-acetylglucosamine)2, β-D-mannose, (α-D-mannose)2. The sugar monomers were then renamed to be recognized by the molecular simulation program, Chemistry at HARvard Macromolecular Mechanics (CHARMM)23 and VMD.15 N-acetylglucosamine (NAG) was renamed to BGLN, β-D-mannose to BMAN, and α-D-mannose to AMAN. Using the Build Structure feature in UCSF Chimera, the glycans were attached at their respective glycosylation sites: N111, N161, N230, N337, and N371. Bonds were added between N’s Nδ2 atom and NAG’s C1 atom. The O1 and HO1 atoms on NAG and HD21 atom on N were deleted.
Solvation and ionization
Aside from a single hydroxide ion present in the active site of the model, no other water molecules were present. Using the Dowser computer algorithm,24 42 internal water molecules were added to the protein structure. Next, the program Solvate25 placed 13,593 rigid TIP3 water molecules around the protein with a minimal solute-boundary distance in a 10Å thick water shell and generated disordered water molecules to mimic the true nature of water. An additional 12,708 external water molecules were added using VMD’s Solvate plugin to form a proper cubic shape around the protein structure to satisfy periodic boundary conditions. In total, 26,301 water molecules were placed in and around TyrD. After the addition of water molecules, VMD’s AutoIonize plugin was used to calculate the system’s net charge and add 13 sodium ions to neutralize the system.
Molecular dynamics
To generate a protein structure file (psf), two tcl scripts were written. The first called for a separation of each segment of TyrD to be written into individual pdb files. The second script used CHARMM topology files to identify each atom of the structure, apply patches, and reconnect the individual segments. The script aliased (renamed) several residue and atom names to be recognizable by the CHARMM force field and applied important patches to the system: two disulfide bonds and several patches pertaining to the glycans. In order to more easily find the glycan atoms in need of patching, a script (a gift from Tristan Croll, Queensland University of Technology, Australia) was used to generate a list of glycan patches. CHARMM version 36 was used for the protein, carbohydrate, and glycopeptide topology/parameter information. The water and ions stream file customized for NAMD was used for water molecules and the hydroxide ion.
In addition, copper parameters were extracted from CHARMM 22 metal topology and parameter files and then added to both the protein topology and parameter files. Custom parameters were added for the coordinate covalent bond found between the coppers and their respective Nε2 atoms. The Van–der-Waals radii, force constants, and bond and angle measurements were taken from the PM0079416 model.19 Additional improper and dihedral values were derived from Ungar et al.26 Lastly, TyrD’s psf file was edited to include bond, angle, dihedral, and improper information for the copper active site.
The minimization and equilibration of the TyrD structure was performed using several different methods. First, TyrD was minimized for 20,000 1fs steps at a temperature of 300K, with no temperature or pressure control using NAMD Scalable Molecular Dynamics,27 where rigid bonds were set to water only, periodic boundary conditions were used to maintain proper volume fluctuations and the cubic shape of the system, and particle mesh Ewald (PME) grid spacing was set to 1.0Å. Once complete, the system was verified as fully minimized by graphing the output of total energy, bond energy, and concentration gradient from the log file. Second, TyrD was equilibrated using the velocities, coordinates, and dimensions files created during previous minimization runs, by slow heating from 100K up to 310K over 5,000 steps. Finally, after 5,000 steps, the system was equilibrated in an isothermal-isobaric (NPT) ensemble, with temperature and pressure held constant using Langevin dynamics. Using the NIH’s Biowulf computer cluster,28 a total of 250ns of molecular dynamics production runs was obtained.
Secondary structure evaluation was also used to measure TyrD’s improvement over molecular dynamics simulations. Initial secondary structure prediction was obtained for TyrD’s amino acid sequence using the PHD Secondary Structure Prediction Method.29 The actual secondary structure characteristics were then calculated for the initial, 54ns, 122ns and 250ns model frames using NIH’s Helix System’s StrucTools server.30 The predicted and actual secondary structure characteristics were then compared.
T373K mutant variant creation
The T373K missense mutation causes a severe form of albinism, OCA1A. Therefore, we paid a special attention to the modeling of this mutant. The T373K mutant homology model was created as follows: in the TyrD model, T373 was mutated to lysine and N371 was deglycosylated and mutated to aspartic acid, to mimic experimental conditions for the mutant variant. Identical force field parameters were used except for deleted patches for the missing glycan at residue N371. The model of the T373K mutant was solvated using the same software and parameters, and 13 sodium ions were added to create a neutral environment. Next, T373K underwent the exact same molecular dynamics simulations as TyrD. Local energy minimization was confirmed before equilibration simulations began. Molecular dynamics production runs of 250ns were achieved to accurately compare T373K with TyrD.
Free energy analysis
First, specific frames in the NAMD trajectory files were chosen based on RMSD data and total energy. The selection of individual nanosecond frames was based on the appearance of equilibrium in the RMSD graph and a correlation to a low total energy point. Gibbs free energy changes for each mutation were analyzed using the semi-empirical method of evaluating protein stability from the atomic structure of protein, FoldX.31 FoldX command files were written to first repair the pdb files, then calculate free energy changes for the specified mutations. To repair the 54ns and 122ns TyrD models, the “RepairPDB” command was implemented in a FoldX configuration file. Next, configuration files were written to run the “BuildModel” command. The missense mutations (T373K/N371D, R402Q, P406L, R422Q, and R422W) were listed in a separate text file. The pdb files were then restored to pre-CHARMM format for compatibility with the program. The following individual mutations were analyzed: T373K, P406L, R402Q, R422Q, and R422W. Free energy changes were calculated as a difference between protein stabilization energies of each mutation and the TyrD protein. In addition, OCA1 mutations obtained from the Albinism Database and previous clinical studies were analyzed.
Small molecule docking
Using Schrodinger’s Glide software,32–34 the binding affinity and possible binding positions of tyrosinase activators and inhibitors were calculated. The small molecule compounds docked to tyrosinase were L-tyrosine, L-DOPA, 3-HAA, and kojic acid. Metal coordination restraints were placed so that a phenol oxygen atom on the small molecules would interact (within 3Å) with one or both copper ions. Docking was performed both with and without a water molecule in the active site to compete with the copper ions and determine any effects on small molecule binding. Additionally, kojic acid was docked along with tyrosine to see how the inhibition effect may work.
Experimental urea-induced equilibrium unfolding/refolding
Proteins TyrD and OCA1B-related mutant variants, R402Q, R422Q, R422W, and P406L were expressed and purified as previously described.1 Protein unfolding/refolding experiments were performed as described in.2 Briefly, these experiments used intrinsic tryptophan fluorescence, measured at an excitation wavelength of 285nm. Emission spectra of TyrD and OCA1B-related mutant variants were recorded in the range between 300 and 400nm, using a SpectraMax i3 microplate reader (Molecular Devices, CA). Proteins (10μM) were in 10 mM phosphate buffer, pH 7.4 with different concentrations of urea (from 0 to 8M, at 0.2M increments). For refolding, proteins were dialyzed against 10mM phosphate buffer, pH 7.4 for 24h. Unfolding and refolding curves were measured as a 360/320nm ratio. The experimental curves were fit to a Boltzmann function in Origin Lab.
Atomic model of human tyrosinase
The homology model of TyrD and the structure of the tyrosinase active site are shown in Figure 2. Protein backbone is shown as a pink, new cartoon, ribbon (Figure 2A). The two copper ions are depicted by orange spheres and located in the center of the active site. Six total glycans and glycosylation sites are depicted in a white, ball-and-stick mode. TyrD is solvated and neutralized with sodium ions. Sodium ions are shown as blue spheres. Residues 106-451 were modeled in a structure. In the copper center active site, CuA is coordinated by covalent bonds to Nε on histidines 180, 202, and 211 (Figure 2B). CuB is coordinated by covalent bonds to Nε on histidines 363, 367, and 390. An OH- ion bridges CuA and CuBThe central portion of the tyrosinase domain is a four α–helical bundle structure, which maintains an active site containing two copper atoms, CuA and CuB. The locations of the copper atoms within the active site are coordinated by histidines 180, 202, and 211 (CuA) and histidines 363, 367, and 390 (CuB) and are highly conserved amongst tyrosinase species.
The OH- ion was placed as a bridge between CuA and CuB to reflect experimental studies depicting the likelihood of a hydroxide ion present in Tyr’s met form. The distances between the copper atoms and their coordinating Nε2 nitrogens on histidine’s imidazole ring were measured. CuA was measured to be approximately 2.0Å in distance from the Nε atom on histidines 180, 202, and 211. CuB was also approximately 2.0Å from the Nε atom of histidines 363, 367, and 390. The quality of the model was verified using secondary structure analysis. The analysis results were performed at different times of 250ns of molecular dynamics and are shown in Table 1. The initial protein model of TyrD was incomplete and it therefore has a low β-sheet structure percentage. However, α-helix percentages (which were comprised primarily of the helical bundles forming the active site) were initially higher (40%) but decreased and quickly matched the predicted sequence values (30.9%) close to that was previously observed using circular dichroism.1
Measurements |
Predicted29 |
Initial TyrD Model |
54ns TyrD Model |
122ns TyrD Model |
250ns TyrD Model |
250ns T373K Mutant |
Total Amino Acids |
346 |
346 |
346 |
346 |
346 |
346 |
β-Sheet Residues |
55 |
4 |
4 |
10 |
10 |
12 |
Percent β-Sheets |
15.90% |
1.16% |
1.16% |
2.90% |
2.90% |
3.50% |
α-Helix Residues |
106 |
138 |
107 |
107 |
106 |
112 |
Percent α-Helices |
30.64% |
40% |
30.9% |
30.9% |
30.6% |
32.4% |
Table 1 Secondary structure analysis. A secondary structure comparison of predicted, initial, 54ns, 122ns and 250ns TyrD models is shown
Overall structure quality and local errors for 54ns and 122ns TyrD were evaluated using the Swiss Model QMEAN Server.35 The TyrD 54ns and 122ns Z-scores and QMEAN residue scores demonstrated that while all-atom interactions and torsion energies improved for 122ns versus 54ns, all other areas either remained constant or slightly decreased in reliability.
Protein glycosylation
In the TyrD model, potential glycosylation sites1 with exception of Asn 86 were modified by the addition of the most biologically common eukaryotic core glycan to the asparagine side chain, as described in the Methods section. We then analyzed the effects of glycosylation on TyrD by calculating the accessible surface area and compared the results of this analysis with our experimental data. In our simulations, the surface area did not change after 100ns. Therefore, we uploaded both our initial TyrD model and the model after 100ns to YASARA. We first calculated the solvent-accessible surface area of each structure, then deleted one glycan at a time, measuring the change in surface area (Table 2). Calculations were repeated for the initial and 100ns models. With molecular dynamics refinement, the sites with the greatest surface area losses became N290, N371, and N337. In addition, N290, N371, and N337 almost doubled in surface area loss compared with the largest amounts previously calculated for the initial structure.
De-Glycosylation Site |
Surface Area Å2 |
Difference Å2 |
Surface Area Å2 |
Difference Å2 |
Fully Glycosylated |
19,056.30 |
0 |
23,442.60 |
0 |
N 111 |
18,227.30 |
-829.00 |
22,659.50 |
-783.10 |
N 161 |
18,268.27 |
-788.03 |
22,970.22 |
-472.38 |
N 230 |
18,299.89 |
-756.41 |
22,653.15 |
-789.45 |
N 290 |
18,525.51 |
-530.79 |
22,311.48 |
-1,131.12 |
N 337 |
18,325.68 |
-730.62 |
21,952.56 |
-1,490.04 |
N 371 |
18,611.01 |
-445.29 |
22,032.36 |
-1,410.24 |
Table 2 Solvent-accessible surface area for the initial and 100ns TyrD models
Molecular modeling of mutant variants
Atomic models of disease-related mutant variants T373K, R402Q, R406L, R422Q, and R422W studied biochemically previously1,2 were generated to understand the consequences of the changes to protein structure. All variants were superimposed over the TyrD structure for further comparison (Figure 3). OCA1A mutation T373K is closest to the active site. OCA1B mutations R402Q, P406L, R422W, and R422Q are exposed at the protein surface. In Figure 3 the TyrD amino acid side chain is shown in baby blue and the mutant amino acid side chain is shown in tan. Panel A shows T373K/N371D–at residue 373, lysine is superimposed over threonine and at residue 371, aspartic acid is superimposed over asparagine; both mutations occur simultaneously. Panel B shows mutation P406L with leucine is superimposed over proline at residue 406. Panel C demonstrates mutation R402Q, where glutamine is superimposed over arginine and residue 402. Panel D shows mutation R422Q with glutamine is superimposed over arginine at residue 422. Panel E demonstrates R422W, where tryptophan is superimposed over arginine and residue 422.
The T373K mutation is located at the N371 sequon and is important for protein stability.3 This mutation was therefore chosen for equilibration using 250ns molecular dynamics in water. The final model of T373K at 250ns was compared with the TyrD model in 250ns simulations. T373K was first superimposed over TyrD to observe structural changes in the native protein conformation (Figure 4). Immediately noticeable was the difference in side chain locations. Residue 373, now lysine in the T373K mutant, was pointed outwards, away from the active site, while threonine 373 in TyrD was pointed inwards, towards the copper center active site. In the T373K mutant, a 3-10 helix began to form between residues 372 and 375. In addition, the secondary structure was analyzed on NIH’s Helix System’s StrucTools,30 with the results showing an increase in both alpha helices and beta strands (Table 1). Interestingly, an increase was noticeable in the helices near residue 371. The Figure 4 shows key structural changes in both structures. TyrD’s ribbon is colored orange, while T373K’s ribbon is depicted in pink. Labeled residues are new helical formations in the T373K mutant. The insert shows the conservation of a β–sheet formation between the N and C termini in both TyrD and the mutant.
Trajectory files (250ns) for both TyrD and T373K were uploaded into VMD. RMSD for both models were calculated and compared at the Figure 5. QMEAN score was evaluated on the QMEAN server36 and is a composite score consisting of a linear combination of six terms. Pseudo-energies of the contributing terms are given together with their Z-scores, with respect to scores obtained for high-resolution experimental structures of similar size solved by X-ray crystallography. A graph of the RMSD comparison for TyrD at 54ns is shown in Figure 5A. Figure 5B shows overall Z-scores for TyrD at 122ns. In addition, a heatmap was created to show RMSD changes per amino acid residue (Figure 5C). Total error (in angstroms) for each amino acid residue in TyrD. TyrD at 54ns (seen in blue) is compared to TyrD at 122ns (seen in red). While the RMSD for each protein model shows, less overall deviation in the T373K mutant, a comparison between the T373K and TyrD heatmaps shows that T373K has many more focused areas of deviation from the original structure than TyrD. Additionally, both copper active site regions in T373K began to deviate greatly, showing very little conservation over 250ns. Lastly, while the region between residues 245 and 305 in TyrD showed deviations mostly ranging from 3 to 8Å (with 3 residues–110, 198, and 355–jumping to almost 20Å), the same region in T373K consistently deviated between 3 and 15Å.
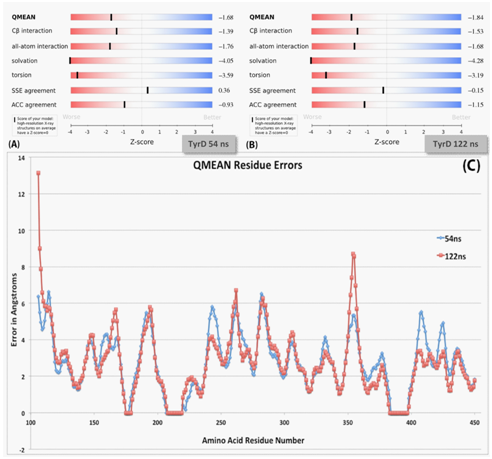
Figure 5 QMEAN Z-scores of TyrD at 54ns and 122ns.35
Mutant variants and protein stability changes
The mutant variant’s atomic models were used for the computational analysis of protein stability and comparison of these data with the free energy changes ΔΔG obtained experimentally as briefly described in Methods section.2 ΔΔG were calculated for the individual mutations: R402Q, P406L, R422Q, and R422W. Both calculated and observed values were plotted on a scatter plot and are shown in Figure 6. Free energy changes were calculated for the 54ns and 122ns TyrD models. 54ns ΔΔG values are shown in purple, while 122ns ΔΔG values are shown in orange. Mutation sites are labeled. Computational free energy changes, ∆∆Gcalc, are shown on the y-axis with experimental values, ∆∆Gobs, on the x-axis.
In our studies, we have an interest in the direct effects of specific mutations on Tyr activity. For this purpose, we gathered clinical data from three published studies of OCA patients with sequenced DNA and a clinical, phenotype diagnosis.10,12,13 The patients were geographically diverse, with one study focusing on Chinese OCA patients,12 one focusing on Iranian OCA patients,13 and another documenting the ethnic heritage of each patients’ parents.10 Using FoldX and our 54ns TyrD model, we obtained ΔΔG values for all documented mutations for the 107 patients to determine the effect of OCA1 mutations on tyrosinase’s protein stability (Table 3). Mutations categorized as OCA1A or OCA1B were obtained from the University of Minnesota’s Albinism Database (www.ifpcs.org/albinism) and were already classified with an OCA1A or OCA1B molecular diagnosis. OCA1 mutations marked with an asterisk were obtained from DNA sequenced from patients treated in the National Institutes of Health Clinical Center. Mutations marked as “additional” were obtained from the three previously mentioned studies and did not appear in the Albinism Database.10,12,13 The S192Y and R402Q mutations are known polymorphisms and were classified as such. We found that 14 mutations could cause an additional stabilization of the protein structure in comparison to that of wild-type TyrD (ΔΔG <0). For other mutations associated with protein unfolding (ΔΔG >2kcal/mol), 29 mutations are associated with OCA1A, with only four associated with the OCA1B phenotype. This suggests that full or partial loss of tyrosinase’s stability might be associated with the cause of OCA1 albinism.
Mutation |
ΔΔG, kcal/mol |
Type |
Mutation |
ΔΔG, kcal/mol |
Type |
Mutation |
ΔΔG, kcal/mol |
Type |
G106R |
-1.15 |
OCA1A |
P313R |
14.36 |
OCA1A |
G436R |
-1.67 |
OCA1A |
G109R |
1.56 |
OCA1A |
E328Q |
1.29 |
OCA1A |
F439V |
1.13 |
OCA1A |
I123T |
4.03 |
OCA1A |
S339G |
2.97 |
OCA1A |
G446S |
2.94 |
OCA1A |
Y149C |
1.5 |
OCA1A |
F340L |
2.76 |
OCA1A |
Y449C |
5.36 |
OCA1A |
P152S |
2.92 |
OCA1A |
G346E |
14.25 |
OCA1A |
K142M |
-2.28 |
OCA1B |
F176I |
3.62 |
OCA1A |
A355E |
0.45 |
OCA1A |
H202R |
-0.33 |
OCA1B |
Y181C |
1.06 |
OCA1A |
A355P |
0.84 |
OCA1A |
P209R |
5.63 |
OCA1B |
P205T |
3.42 |
OCA1A |
Q359L |
0.43 |
OCA1A |
V275F |
0.5 |
OCA1B |
A206T |
0.56 |
OCA1A |
S360G |
-0.66 |
OCA1A |
R299S |
3.73 |
OCA1B |
R212K |
2.56 |
OCA1A |
S361R |
1.64 |
OCA1A |
T325A |
-0.19 |
OCA1B |
L216M |
0.54 |
OCA1A |
H367R |
-0.61 |
OCA1A |
Y327C |
3.35 |
OCA1B |
R217W |
1.26 |
OCA1A |
Y369C |
0.51 |
OCA1A |
M332T |
0.97 |
OCA1B |
R217Q |
1.42 |
OCA1A |
M370T |
1.67 |
OCA1A |
N371Y |
-0.43 |
OCA1B |
R217G |
3.33 |
OCA1A |
N371T |
2.98 |
OCA1A |
R402G |
1.23 |
OCA1B |
E219K |
2.6 |
OCA1A |
G372R |
1.22 |
OCA1A |
P406L |
1.03 |
OCA1B |
E221K |
-1.47 |
OCA1A |
T373K |
1.66 |
OCA1A |
R422Q |
0.48 |
OCA1B |
W236S |
4.17 |
OCA1A |
T373K, N371D |
3.89 |
OCA1A |
R422W |
0.14 |
OCA1B |
R239W |
0.06 |
OCA1A |
S380P |
2.05 |
OCA1A |
Y433C |
0.19 |
OCA1B |
G253R |
-0.11 |
OCA1A |
N382K |
0.04 |
OCA1A |
D448N |
2.08 |
OCA1B |
G253E |
2.38 |
OCA1A |
D383N |
0.04 |
OCA1A |
T373K, N371D, W272C |
2.96 |
OCA1* |
H256Y |
-2.28 |
OCA1A |
H390D |
1.62 |
OCA1A |
P406L, P431T |
3.95 |
OCA1* |
W272R |
0.25 |
OCA1A |
A391E |
7.12 |
OCA1A |
H180R |
0.15 |
Additional |
W272C |
0.63 |
OCA1A |
V393F |
-0.63 |
OCA1A |
H202Q |
0.31 |
Additional |
L288S |
1.01 |
OCA1A |
S395N |
1.73 |
OCA1A |
H211R |
4.66 |
Additional |
C289G |
4.23 |
OCA1A |
W400L |
2.95 |
OCA1A |
Y235H |
3.62 |
Additional |
C289Y |
4.76 |
OCA1A |
R403S |
1.25 |
OCA1A |
W236R |
2.94 |
Additional |
C289R |
5.19 |
OCA1A |
H404P |
0.61 |
OCA1A |
D249G |
0.7 |
Additional |
E294K |
-0.33 |
OCA1A |
P412A |
2.56 |
OCA1A |
R299C |
3.61 |
Additional |
E294G |
0.87 |
OCA1A |
G419R |
11.7 |
OCA1A |
M332I |
0.62 |
Additional |
R299H |
3.06 |
OCA1A |
P431L |
2.2 |
OCA1A |
S192Y |
-0.35 |
Polymorphism |
D305E |
0.01 |
OCA1A |
P431T |
2.9 |
OCA1A |
R402Q |
0.76 |
Poly/OCA1B |
L312V |
2.1 |
OCA1A |
Table 3 Free energy changes, ΔΔG, calculated for known OCA1 clinical mutations
Activator and inhibitor docking
Four small-molecule compounds were docked into the active site of the 54ns TyrD structure to validate the applicability of the TyrD model in a drug-search related study. The compounds were docked both with and without the presence of a water molecule at the active site. For each docking simulation, the binding affinity (GlideScore) was calculated and the distances between key atoms were measured. When docking studies were performed on L-tyrosine (Figure 7A), we observed what has been shown to occur in experimental studies with bacterial Tyr: the phenol group bridges and coordinates with the copper ions, likely picking up an oxygen atom being held in place by CuA.37 Tyrosine is docked in the correct position when a water molecule is present in the active site and able to interact with one of the copper ions. Tyrosine’s phenol group is coordinated with the coppers and the binding affinity (GlideScore) is calculated at -6.74kcal/mol. However, the docking results place the tyrosine hydroxyl group at 4.8Å from Cu-A. When a water molecule is competing for coordination with a copper ion, L-DOPA is slightly rotated and the binding affinity decreases (Figure 7B). Figure 7C: when 3-HAA’s carboxyl group is fully deprotonated, 3-HAA achieves its best pose and both oxygen atoms are coordinated with the copper ions (within 3.5Å). Kojic acid was first docked in the active site and tyrosine was docked second (Figure 7D).
We have developed a homology model of glycosylated human TyrD. Our model is attributed to the use of several strong crystal templates from various species and the incorporation of a fragmented, human active site previously published.19 One of these important crystal structure templates was derived from octopus hemocyanin, which was similarly used in 200738 and 2010,39 in inhibitor drug docking studies. We further improved quality by using several homology modeling servers and computer programs, before selecting the best overall model. Finally, we carefully avoided any models that produced backbone perturbations in the active site. A common homology model seen in previous studies shows a backbone loop segment inserted between the two copper atoms39,40 – a highly unlikely conformation. As previously demonstrated in other studies, CuA and CuB both play integral roles in monophenolase and diphenol oxidase activity, regulating the binding of L-tyrosine, L-DOPA, and L-dopaquinone, which are all essential steps to producing melanin.41,42 Further studies have highlighted the likelihood of an oxygen atom bridging the two copper atoms, which may initially come from hydroxide or water.19,38 These revelations are essential in predicting Tyr activators and inhibitors and make a properly designed active site extremely important. In addition, we have shown the importance of using molecular dynamics as a method of refinement and analysis. While other studies have created active site models with energy minimization40 or full models with 20ns or less of molecular dynamics simulations,43 we achieved 250ns for both our TyrD and T373K mutant models. Our Z-scores also showed a significant improvement when compared to models created in previous studies. Through molecular dynamics simulations, we have further refined our protein in terms of total energy, secondary structure, and active site behavior. Our findings provide some insight into OCA1 variability and pave the way for future drug design.
The accuracy of homology modeling relies on the availability of protein crystal structures. The use of molecular dynamics has enabled us to increase the accuracy of homology modeling and create a TyrD structure to use for successful mutation analysis and free energy calculations. As shown in Table 1, we achieved a realistic percentage of α-helices in our secondary structure. We also observed some improvement in the formation of a β-structure between the N- and C-terminal amino acids. Without a longer molecular dynamics simulation however, we are unable to better improve the formation of β-strands. Interestingly, in the T373K mutant, a higher percentage of α-helices and β-strands were observed, especially surrounding the active site. As described in previous studies,7,8 the loss of the glycan at residue 371 seems to increase protein misfolding. In OCA1A, Tyr is degraded in the ER because of improper folding.9 In the T373K mutant, studies have shown that the loss of the glycan at N371 prevents Tyr from leaving the ER, thus creating an OCA1A disease phenotype.7,8 In OCA1B, Tyr can move forward in its normal path, but specific activity is reduced.1
The glycosylation site at N371 is essential for normal enzymatic activity.8 Surface area calculations show the structural differences between the TyrD and the T373K mutant, as well as the significant effect of the loss of glycan 371 (Table 2). Glycosylation site 337 and 371 both exhibited large decreases in solvent-accessible surface area, indicating a significant change in water interaction compared to that of glycans in other positions. This agrees with our experimental observations showing the importance of these positions for protein stability. With continued molecular dynamics studies, we hope to better identify the structural effects caused by the loss of glycan 371.
Using our refined TyrD model, we have found the Gibbs free energy changes of clinically significant mutations. A correlation was found between the experimental and computational ΔΔG values for the TyrD and mutant variants. For the 54ns TyrD model, the R2 linear correlation is 0.91, while the Pearson’s R is 0.95. For the 122ns TyrD model, the R2 linear correlation is 0.78176, while the Pearson’s R is 0.88417. Both indicated a strong similarity between the computational and experimental ΔΔG. The ΔΔG values for R402Q, P406L, R422Q, and R422W of the TyrD 54ns model are as follows: 0.45, 1.04, 0.55, and 0.27kcal/mol, respectively. The ΔΔG values for the TyrD 122ns model are: 0.33, 1.87, 0.93, and 0.50kcal/mol, respectively. However, the absolute ΔΔG values for 54ns more closely matched the experimental ΔΔG range. The speculation of the severity of the T373K mutation comes from the disruption of glycosylation at residue 371. Without the N-X-S/T glycosylation sequon (where X is any amino acid except for proline), a glycan cannot properly be attached to the polypeptide chain. The T373K mutation is often seen in patients diagnosed with OCA1A, indicating a complete loss of enzymatic activity. In contrast, our T373K/N371D mutant values could not be accurately compared with the experimental ΔΔG values because the protein was found to be completely inactive and degraded after purification.2 Moreover, our computational ΔΔG value may also be unreliable owing to FoldX not recognizing, or being able to account for, the glycan atoms at position 371.
Over the course of MD simulations, the active site remained conserved, with CuA and CuB maintaining their coordinate bonds with their respective histidines. Also, the hydroxide ion, held in place only by electrostatic potentials, moved away from bridging the two copper ions. However, numerous water molecules moved in and out of the active site, ensuring that an oxygen atom was constantly bridging CuA and CuB. To understand the possible implications of the TyrD model in drug design, we performed a protein docking with tyrosinase substrates, L-tyrosine and L-DOPA, tyrosinase inhibitor kojic acid, and protein activator 3-HAA (Figure 7). In a docking of L-tyrosine shown in Figure 7A the most highly scored position occurred with the phenol oxygen protonated (Glide Score ~-6.7kcal/mol). In addition, removing an active site water molecule tested the importance of an external oxygen atom. This caused L-tyrosine to “flip” and use its carboxyl oxygen atoms for coordination instead. We also performed similar tests with the L-DOPA compound and found that the most effective docking pose occurred when no free oxygens (e.g. water or hydroxide) were coordinating with the copper ions (Figure 7B). Moreover, the best L-DOPA pose occurred when the phenol oxygens were both protonated and coordinated with CuA and CuB within 3Å. When L-DOPA’s phenol oxygens were held by double bonds, L-DOPA’s Glide Score decreased from approximately -6.0kcal/mol to -3.2kcal/mol and the molecule was pushed further away from the copper active site (>3Å). This suggests that oxidation of the phenol groups is necessary to release L-DOPA from Tyr’s active site. 3-Hydroxyantrnilic acid (3-HAA) is an activator of Tyr catalytic activity,44 while kojic acid is a well-known inhibitor of Tyr.45,46 Like L-DOPA docking, 3-HAA showed an improved binding affinity to both coppers within the active site when a competing water molecule was not present (Figure 7C). In addition, 3-HAA was able to position itself closest to the copper ions when both phenol oxygens were deprotonated, and double bonds were alternating (data not shown). The distance and GlideScore binding affinity both decreased in the presence of hydrogens. Kojic acid, when docked alone, had less desirable GlideScores and positioned itself with its ring closest to the copper ions (Figure 7D). The presence or absence of water did not alter the GlideScore or the kojic acid positioning. When paired with tyrosine, kojic acid docked into the coordinating position with the two copper ions first, followed by tyrosine docking second with a distance as 10Å to the copper ions. Our analysis demonstrated that the TyrD model could be useful for an in-silico search of tyrosinase small molecules inhibiting or activating the catalytic function of tyrosinase.
Although a phenotype of white hair and white skin pigment at birth usually correlates to a diagnosis of OCA1A, diagnosis may change depending on DNA analysis, as well as the possible development of pigment later in life. Gene sequencing of the TYR gene often reveals an inherited mutation on two alleles, one from the maternal side and one from the paternal side. New studies continue to arise regarding heterozygous and single mutation OCA patients, making an analysis such as ours even more important for predicting phenotype. Birth prevalence and mutation spectrum in Danish patients with autosomal recessive albinism was obtained by sequencing of all OCA-related genes in OCA patients, though several patients were identified with a single or no mutation. Addressing both TYR alleles and all mutation types does key to a better understand OCA1. However, much research is still devoted to the possibility of currently unknown non-TYR genes contributing to Tyr deficiency and/or OCA1, including the TYR-like pseudo gene.
Our findings help to further clarify the role of Tyr in OCA1. Our TyrD model paves the way for further work involving glycosylation analysis, mutation analysis, and drug design. Our research is limited however, by available force fields for copper and other metals. Despite recent gains in quantum mechanics, metalloproteins remain some of the most difficult proteins to work with computationally. Copper’s number of valence electrons varies and is especially difficult to accurately represent when it shifts from Cu+2 to Cu+1 during the Tyr enzyme reaction. Our model is based on Tyr in its met, native form, as it exists just before its enzyme reaction occurs. In this state, the two active site copper ions exist as Cu+2. For small molecule binding and drug-docking studies, further refinement of the active site force field may be required. Lastly, our research was limited by our use of a truncated model and relatively short molecular dynamics simulations, which hinders a full understanding of the role of protein folding in Tyr. Significantly longer molecular dynamics simulations are required to visualize protein folding in understanding the relationship between Tyr’s structure and function. Finally, our results could be used to understand the role of mutant variants in melanin pigment production, in silico searches for inhibitors and activators of tyrosinase activity, and genotype-to-phenotype analysis of OCA1.
This research was supported by the Intramural Research Program of the NIH. This work utilized the computationalresourcesof the NIH HPC Biowulf cluster (http://hpc.nih.gov). We also are thankful for the use of commercial molecular modeling software Maestro on the network through the Center of Molecular Modeling, CIT/NIH. We thank Mike Blaber at Florida State University for his suggestion on the structure of the most biologically common eukaryotic core glycan. We also thank Kenneth Young II from our Lab for a critical reading of the manuscript.
The author declares that there is no conflict of interest.
This research was supported by the Intramural Research Program at the National Eye Institute of the NIH, ZIA EY000476-09 to Y.V.S.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.