eISSN: 2574-8092


Research Article Volume 8 Issue 3
Cuban Neuroscience Center, Cuba
Correspondence: Eduardo Garea-Llano, Cuban Neuroscience Center, Cuba
Received: November 25, 2022 | Published: December 7, 2022
Citation: Garea-Llano E, Martinez-Montes E, Gonzalez-Dalmaus E. Affectation index and severity degree by COVID-19 in Chest X-ray images using artificial intelligence. Int Rob Auto J. 2022;8(3):103-107 DOI: 10.15406/iratj.2022.08.00252
The Covid-19 pandemic has caused the congestion of intensive therapies making it impossible for each to have a full-time radiology service. An indicator is necessary to allow intensivists to evaluate the evolution of patients in advanced state of the disease depending on the degree of involvement of their lungs and their severity in chest X-ray images (CXR). We propose an algorithm to grade the affectation of lungs in CXR images in patients diagnosed with COVID-19 in advanced state of the disease. The algorithm combines the assessment of image quality, digital image processing and deep learning for segmentation of the lung tissues and their classification. The proposed segmentation method is capable of dealing with the problem of diffuse lung borders in CXR images of patients with COVID-19 severe or critical. The calculation of the affectation index (IAF) consists of the classification of the segmented image by establishing the relationship between the number of pixels of each class. The IAF index of lung affectation in CXR images and the algorithm for its calculation. A correlation was established between the IAF and the international classification of the degree of severity established by radiologists.
Keywords: machine learning, supervised classification, COVID-19, x-ray images, digital image processing
After more than two years in the fight against the COVID-19 pandemic, the protocols and forms of action in the treatment of patients in serious or critical condition have been constantly renewed in order to arrive in the first instance to save the lives of these patients.
The large number of patients who arrive at the intensive care services has caused in many cases the saturation of these services, this has made it practically impossible for each of them to have a full-time specialist in radiology, for this reason it is necessary have an indicator that allows intensivists to automatically evaluate the evolution of patients based on the level of involvement of their lungs.
Batista et al.,1 carry out an analysis of the role of images in the diagnosis of pneumonia caused by COVID-19. The authors state that chest X-rays have proven effective in the early detection and management of pulmonary manifestations caused by COVID-19. Recent studies show that signals on chest X-ray images that are characteristic of people infected with COVID-192,3 may decrease or increase over time depending on the state of severity of the patient and the effectiveness of the applied protocols.4,5 In the bibliographic review carried out5-8 some works were found that propose the evaluation of the degree of severity based on radiological classifications of lesions and their severity from the use of deep learning-based image segmentation methods.8
The quality of a medical image is determined by its capture method, the characteristics of the equipment and the image variables selected by the operator. Image quality is not a single factor, but rather a combination of at least five factors: contrast, blur, noise, artifacts, and distortion.9
Of the variety of factors that negatively influence the quality of the images, the degree of sharpness is one of the most important, since if the image is blurred or out of focus, the details of the texture of the lungs could be lost. , that is, of the internal structures of the same, which are those that can suffer the effects of the disease.10
In Garea-Llano et al.,11 an analysis of the use of the Kang and Park filter was performed using images of eyes taken in the near infrared spectrum. The kernel is capable of filtering high frequencies within the image texture better than other state-of-the-art operators, in addition to having a low computational cost due to its reduced size. On the other hand, the entropy of an image has proven to be a good indicator of the volume of information contained in it.12 Entropy only depends on the volume of gray levels and the frequency of each level.
The fundamental objective of the lung segmentation process in the CXR image is to be able to concentrate the classification process only in the lung regions, avoiding interference in the process that may be caused by the rest of the image regions.
Automatic segmentation of CXRs has been extensively studied since the 1970s, at least for segmentation of the lungs, rib cage, heart, and clavicles.13 Conventional methods rely on prior knowledge14 to delineate anatomical objects from CXR images. Modern approaches use deep convolutional networks and have shown superior performance.15 Despite the great advances achieved in the automatic segmentation of these organs, limitations still persist, such as the need to use small-sized CXRs or the irregularity and imprecision of the edges resulting from segmentation that reduce its applicability in clinical settings.
Additionally, CXR images of COVID-19 patients in an advanced stage of the disease make the segmentation process a challenging task because the characteristic conditions of this disease cause whitening of the lung region, which confuses most of the trained algorithms for this task.16
In most of the most recent literature on the use of artificial intelligence in the fight against COVID-19, image classification is understood as the process of assigning to the image a label related to the presence or not of the disease.
In López-Cabrera, et al.17 a study of the state of the art of the most recent methods used in the identification of COVID-19 from CXR images is carried out. It is evident that the trend is towards the application of techniques through deep neural networks. Most of the results obtained are encouraging since they report very good results that even surpass the radiologists themselves, although they are not convinced.18 The authors of López-Cabrera, et al.,17 delve into the peculiarities of this task and the characteristics of the databases taken for the training of the networks and what are the factors that may be causing biases in them. These factors are fundamentally referred to the amount of data, training samples, metadata, and the characteristics of the sensors. They raise the idea that the use of traditional methods of computer vision and machine learning (machine learning) could lead to models with greater capacity for generalization.
In19 a "Radiographic Assessment of Pulmonary Edema" (RALE) system was proposed as a tool for practical use by chest radiologists, this is a numerical scoring system in which the chest is divided in four quadrants, each of which is scored on a numerical scale according to the percentage extension of affectations such as consolidation and the density of ground glass opacities. The RALE score is calculated by adding the product of the scores from the four quadrants, and can range from 0 to 48.
In this sense, the Cuban Society of Imaging (SCI) collegiately adapted this scale as a tool for the work of Cuban radiologists in the fight against COVID-19.27 This modified scale goes from 0 to 8 points depending on the extent of lung involvement. The score from 0 to 4 points is obtained by calculating the involvement of each lung separately and finally adding them.
Figure 1 presents the general scheme of the proposed algorithm, which consists of 4 main steps: 1) image quality evaluation; 2) image enhancement if necessary; 3) segmentation of the region corresponding to the lungs and 4) classification of the pixels of the resulting image and calculation of the index. The following sections explain each of these steps in detail. In the case of the resulting final image, the pixels in green signify regions classified as healthy+bones and those represented in red as signify regions classified as affected.
Context of application and characteristics of the data:
In order to achieve the best possible approach to the context of practical application of the proposed algorithm, a database of CXR images of 3 hospitals that work in the fight against the COVID-19 pandemic in Havana Cuba. The images were taken in the period from May 2020 to April 2021. The database consists of 654 images of the same number of patients, including 500 images of patients diagnosed with COVID- 19 and 154 images of healthy people. The images were obtained from the digital scan in jpg format of the plates printed on acetate and from the files in dcm format obtained directly from the digital x-ray equipment. For the experimental work, all the images were converted to jpg format and scaled to a size of 480 x 480. This is not a public database and has only been used for research purposes with the consent of the hospital institutions. For this, a process was carried out that included anonymizing the data by the hospital institutions, so the researchers did not have access to the biographical data of the patients.
Image quality assessment
In this paper we propose a method for evaluating the quality of the chest x-ray image that is based on the calculation of a quality index (qindex). This is obtained from the estimation of the sharpness of the image and the level of diversity of the texture present in it, using a combination of the Kang and Park filter and a measure of the entropy of the image. The proposed qindex measure is obtained by equation 1.
(1)
Where kpk is the average value of the pixels of the image obtained as a result of the convolution of the input x-ray image with the Kang and Park filter. tkpk is the estimated kpk threshold to obtain a quality image, in Garea-Llano, et al.11 the authors, based on their experimental results, recommend a threshold=15. ent is the entropy value of the image. tent is the estimated threshold of ent at which it is possible to obtain a quality image.
The entropy or average information of an image can be determined approximately from the histogram of the image.12
For the definition of the entropy threshold (tent) we carried out an experiment for which we created a set of 300 good quality CXR images selected from the criteria of radiology experts.
To determine the minimum value of entropy for a quality image. The frequency distribution of entropy showed that good quality CXR images have an entropy greater than 4, (in a range of 4.3 to 5.6, with X ̅=4.9 and s=0.65),
so we propose assuming this value as the value of tent.
The qindex measure can reach values that depend on the thresholds selected for kpk and ent. In this way, considering the values of the thresholds tkpk=15 obtained experimentally in Garea-Llano, et al.11 and tent=4 obtained experimentally by us, the minimum value of qindex to obtain a quality CXR image will be 1, higher values denote higher quality images and values less than 1 denote low quality images.
After evaluating the quality of the input image, the proposed algorithm establishes a conditional step that allows improving the quality of the input image if it presents a qindex < 1. For this, the adaptive histogram equalization method limited by contrast (CLAHE)20 that has given good results in the improvement of x-ray images.21 This method allows to enhance the image even in regions that are darker or lighter than the majority of the image. After applying the CLAHE method to the images whose qindex is less than 1, its improvement is achieved and the qindex is raised to levels greater than or close to 1 (see results section), which allows continuing the subsequent steps of the proposed algorithm.
Lung segmentation
To carry out the segmentation of the lung region, we propose the application of a convolutional neural network (CNN). For this purpose, the previously trained CNNs are adapted using transfer learning. In this work, inductive learning is performed on UNet-CNN22 to segment lung regions. UNet is based on a CNN architecture, this network has shown high precision in several tasks dedicated to the segmentation of structures in electron microscopy and was recently used successfully to segment lungs in CXR with the use of masks obtained manually.23
For network training, 700 images were taken from the Montgomery County Chest X-ray database (National Library of Medicine, National Institutes of Health, Bethesda, MD, USA)24,25 that has its corresponding segmentation masks ("ground truth"). We also used a set of 300 images corresponding to COVID-19 patients from our database. Their segmentation masks were obtained manually following the methodology established in,24,25 under the supervision of a radiologist. These 300 images were chosen under the criteria of having strong effects from COVID-19 and correspond to patients who were admitted to Intensive Care Units due to the degree of severity of the disease.
Supervised classification of the lung image and calculation of the affectation index
This task could also be categorized as a segmentation task on the "segmented image of the lungs" but in order to differentiate between the two we call it "classification" since it is assigns a label to each pixel of the image corresponding to the class to which it belongs. In our proposal, for the classification of the image pixels we chose a classic machine learning algorithm, the Random Forest. This algorithm creates multiple decision trees and combines them to obtain a prediction. In this algorithm additional randomness is added to the model, as the trees grow, instead of looking for the most important feature when splitting a node, it looks for the best feature among a random subset of features, this results in a wide diversity that usually results in a better model.26
The CXR images are in shades of gray where in healthy patients the anatomical elements that make up this region where the lung mass, rib bones and clavicles are found are differentiated. On the other hand, radiological studies16 of the effects caused by COVID-19 have determined that ground glass opacities and consolidation with or without vascular enlargement, interlobular septal thickening and air bronchogram sign are the most common affectations that are observed in the x-ray images of the pulmonary region. These elements are manifested in the change of tones and texture of the lung region from dark tones for the healthy regions of the lung and lighter tones for the affected regions, with a predominance of the texture called "ground glass". Taking these elements into account in this work as a first approximation, we propose to use the tones of the regions to be classified as features for image classification and their combination with texture descriptors such as first-order statistics and binary local patterns, which We will divide into two classes: healthy lung + bones and affectations.
To train the classifier, two subsets of images of the lung region were taken from our database, previously segmented by the proposed method and classified by two radiology specialists. A first part of 180 images was taken as the training set. The images comprise 90 images of healthy lung and 90 images of lung affected by COVID-19. From this set, 4500 samples (pixels) were randomly taken, 2250 samples of each class. All (100%) of the samples corresponding to the effects caused by COVID-19 were taken from the images of patients with the disease, while 90% of the samples of the remaining class (healthy lung + bones) correspond to images of healthy people and 10% to patients with COVID-19. The remaining images comprising the subset were designated as the test set. Taking into account that the classification of the image is developed in each pixel of the image and that as a result of its application it is possible to quantify the number of pixels belonging to each class (healthy lung + bones and affectations) and establish a relationship between each classified region from the image, we propose the calculation of the affectation index (Iaf) by the following expression (2):
(2)
Where: P1, is the sum of the pixels classified as affected region. P0 is the total sum of the pixels classified as healthy region + bones. The values that the Iaf can reach are between 0 and 1. The closer to zero the value of Iaf is, the less degree of affectation the patient will have. (See example in figure 3)
The proposed algorithm was integrated into 4 basic functions (quality assessment, improvement, segmentation and classification).
For the evaluation of the proposed quality measure (qindex), the 654 images of subset 1 of our database were taken, and their quality was calculated using the qindex index. The results obtained show that in the case of images of healthy people, 37% have poor quality, however, in the images of people with COVID- 19 the opposite occurs with 80% of poor quality images. This may be due to the effect caused by conditions of the type such as ground glass opacities in the images with COVID-19 or the origin of a part of these images that were obtained from the scanning of the plates.
The application of the improvement method to images with qindex <1, showed an increase in the quality of the images and with an increase in the qindex for the images of healthy people (X ̅=0.38, s=0.071) and people with COVID-19. 19 ( X ̅=0.30, s=0.27) respectively. In the case of the images of healthy people, only 5 images had a qindex below 1 but very close to this value; while the images of people with COVID- 19 also increased their quality even in the images that were left with qindex less than 1. These results show the usefulness of the improvement method since with its application it is guaranteed that the images that will go to the classification process have better quality than the original images, which may result in better results in the classification.
For the evaluation of the proposed segmentation step, a subset of images from our database was randomly formed. The selected images were not used in the training set of the network. Each image was manually segmented by two medical specialists to obtain the ideal segmentation (“ground truth”) of their segmentation masks. Due to the complexity of the manual segmentation process and the limited time available to specialists, the subset consisted of 20 images (10 normal and 10 covid-19). As a second step, the 20 images were segmented by the proposed method and the segmentation result of each image was compared with its corresponding ideal segmentation, using the calculation of the E1 error rate proposed in the protocol. NICEI (http://nice1.di.ubi.pt/). This metric estimates the ratio of mismatched pixels between automatic and ideal segmentation, the closer the value of E1 to 0, the better the segmentation result.
In the results, the images corresponding to healthy people reach a lower mean E1 value (X ̅=0.17, s=0.031) than in the images corresponding to patients with COVID-19 (X ̅=0.25, s=0.051 ), this is given by the nature of the affectations caused by the disease that implies the presence of the so-called “ground glass opacities” that can confuse the segmentation method. Figure 2 shows an example of segmentation of an image of a patient with COVID-19.
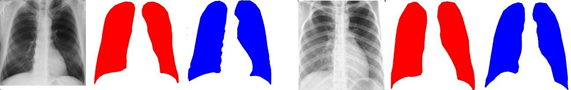
Figure 2 Examples of segmented images of a healthy person (left), COVID-19 patient (right), (in red: “ground truth” mask, in blue: mask resulting from segmentation).
For the evaluation of the proposed classification method, the test set made up of images that were not used in the training set of the classifier was taken: This test set consisted of 77 images of patients with COVID- 19 and 76 of healthy patients. The proposed algorithm was applied to these. Previously, two expert radiologists carried out a manual segmentation and labeling process of the regions affected by COVID-19, which made it possible to count the total number of pixels that make up this class and as pixels corresponding to the healthy lung and bone classes to the rest of the cells. the pixels within the lung region. Automatic classification was applied to these test set images and the Sensitivity and Specificity metrics were calculated, taking into account for each class the correctly classified pixels (TP), those classified as of that class but that really belong to another class. (FP), those classified as from another class but that belong to the evaluated class (FN) and those classified as from other classes and that they really are (TN). The metrics were calculated by the expressions in (3). The average confusion matrices of each class were also obtained.
(3)
A 4-fold cross-validation process was performed for the experiment. The test database was divided into four subsets and the training and cross-testing process was performed. Table 1 shows the confusion matrix, the average of the sensitivity and specificity values and standard deviation (s) obtained. As can be seen in all the results, the sensitivity and specificity values are still high, indicating a good performance of the obtained classifier.
|
Pixels x class |
not |
yes |
Sensibility |
S |
Specificity |
S |
|
healthy class = healthy + bones |
|
|||||
|
No 13683810 |
13215522 |
468288 |
0.964 |
0.012 |
0.966 |
0.018 |
|
Si 10712730 |
382610 |
10330120 |
||||
|
class affectations |
|
|||||
|
No 53612730 |
51200269 |
2412461 |
0.984 |
0.01 |
0.955 |
0.004 |
|
Si 13788810 |
220639 |
13568171 |
|
|
|
|
Table 1 Results obtained in the classification
On the other hand, the presence of pixels falsely classified as affectations is largely due to the similarity of the tones of the bones with the tones that present the true affectations within the lung area (see figure 3). This is a problem that could be solved by applying a bone suppression method to the image, so it is an aspect that we will explore in future research.

Figure 3 Examples of segmented and classified images of a healthy person (left), COVID-19 patient (right), (in red: class affectation, in green: class healthy+bones).
For the study of the correlation between the proposed Iaf and the modified RALE Scoring System, a set of images of patients with COVID-19 (70) and healthy people (30) were selected from our database to whom the automatic classification process in each of the 8 quadrants established for the RALE system. In turn, a group of radiologists, members of the SCI and specialists from hospitals dealing with the disease in Havana, carried out the RALE scoring process of these images.
Figure 4 presents the summary of the results obtained. It can be observed that in 100% of the cases of patients with COVID-19 evaluated, the radiologists classified with value=1 in at least 1 quadrant (presence of affectations) those quadrants where the calculated Iaf the average minimum value was at 0.30 with a standard deviation of 0.02. The Iaf values of healthy people in all quadrants were always below this threshold.
These results show a high correlation between the automatically calculated Iaf value and the RALE classification criteria established to consider a quadrant of the lung region as affected, which allows this threshold (0.30) to be used to automate the classification RALE process from the calculated Iaf by the proposed algorithm.
This paper proposes the Iaf index of lung involvement in CXR images in patients diagnosed with COVID-19. The Iaf is obtained from an algorithm that comprises four fundamental steps that begins with the evaluation of the quality of the images, for which the qindex quality index is proposed, from which an image improvement process is carried out in a manner selective. Next, segmentation of the lung region is performed. With the segmented image, its supervised classification is performed and the Iaf affectation index is calculated.
The results achieved in the experiments carried out on images of healthy patients and those affected by COVID-19 showed high values of sensitivity and specificity in the classification. A correlation was established between the Iaf and the RALE severity degree classification that allows the automation of its estimation as a tool to assist intensivists in decision-making when faced with the impossibility of having a full-time radiology service in intensive therapies. This is a partial result that is still under investigation. Future work is aimed at obtaining a prognostic method for the evolution of severe patients from the combination of the results proposed here with the patient's clinical data. To do this, we explore the use of deep neural networks of the transformer type that allow working with time series.
This work is part of the project "Improving the protocol to deal with COVID-19 using Chest X-ray as a diagnostic and prognostic instrument", financed by the Cuban Center for Neurosciences. We thank the Cuban Society of Imaging and the Hospitals "Luis Díaz Soto" (Naval), Institute of Tropical Medicine "Pedro Kouri" and "Salvador Allende" for their contribution to the conformation of the image databases that allowed us to develop the research.
Author declares there is no conflict of interest.

©2022 Garea-Llano, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.