eISSN: 2379-6367


Research Article Volume 10 Issue 6
1Universidad de Magallanes, Facultad de Ciencias, Chile
2Hospital Dr. Luis Calvo Mackenna, Laboratorio Clínico, Chile
3Universidad de Valparaíso, Facultad de Farmacia, Chile
Correspondence: Alonso De la Rivera Morales, Universidad de Magallanes, Facultad de Ciencias, Punta Arenas, Chile
Received: December 17, 2022 | Published: December 27, 2022
Citation: Morales ADR, Osorio DY, Palma CS, et al. NUDT15, MRP4 and the development of precision medicine in Chile. Pharm Pharmacol Int J. 2022;10(6):230-232. DOI: 10.15406/ppij.2022.10.00392
The genetic structure in Chile is heterogeneous and identifying native, African, European, and Asian components. Increasing the uncertainty about the frequency of genotypic variants and diversifying the response to chemotherapeutic drugs in our population. The individualization of the dose based on Pharmacogenomics is essential to achieve the expected effects of the treatment being more effective and avoiding the toxic effects of drugs such as 6-Mercaptopurine, the main pharmacological agent in acute lymphoblastic leukemia. Whose metabolization is affected by single nucleotide polymorphisms (SNPs) in the enzyme NUDT15 (rs116855232) and the protein MRP4 (rs3765534) related to drug resistance. Both of which cause leukopenia and myelosuppression in patients at standard doses and are not available for detection in Chile. The background of the prevalence of 8.8% in Uruguay and 12.5% in Mexico of the SNP in NUDT15. In addition to international clinical guidelines that recommend reducing the dose by up to 80% depending on the genotype. Is of interest for the development of new techniques in laboratories. In this project both SNPs were implemented using real-time PCR and validated by sequencing being the basis for future studies related to frequencies and their relationship with adverse events. Providing key antecedents for antineoplastic treatment and contributing to the development of a medicine of accuracy in the country.
Keywords: NUDT15, MRP4, pharmacogenomics, 6-Mercaptopurine, precision medicine
In Chile it is estimated that childhood cancer affects 500 new children each year. Most of whom (84.7%) are diagnosed and treated by public health with acute lymphoblastic leukemia (ALL) being the most common.1 It is one of the scenarios where pharmacogenomic studies are very useful because chemotherapeutic drugs have a narrow therapeutic margin, risk of toxicity, and exhibit intra- and inter-patient variability which makes this a field of research of interest for the development of new laboratory methods, capable of providing key clinical information for cancer treatment.2
Pharmacogenomics refers to the study of genes related to the metabolism, transporters, or ion channels of drugs with individual variation to drug response being one of the main problems in achieving treatment efficacy and has been so relevant that applied pharmacogenomics to the clinic has changed the paradigm of how to achieve an effective therapeutic response. In addition to the understanding of how to use genetic biomarkers.3
6-Mercaptopurine
6-Mercaptopurine (6-MP) is the main drug in ALL and exerts its cytotoxic action through thioguanine nucleotides (6-TGN) which are incorporated into DNA and leading the cell to apoptosis. This action is the cause of the main adverse effects of this drug.4 Its metabolization consists of the conversion of 6-MP to its active metabolite 6-TGN, where the enzyme NUDT15 degrades the phosphorylation of the nucleotide thus preventing its entry and the enzyme thiopurine methyl transferase (TPMT) produces inactive methylated metabolites that reduce the formation of 6 -TGN (Figure 1).5 On the other hand, MRP4 is a protein that expels cytotoxic drugs such as 6-TGN from the cell, it is expressed in hematopoietic cells and hematological toxicity has been related to low levels of expression of this protein.6
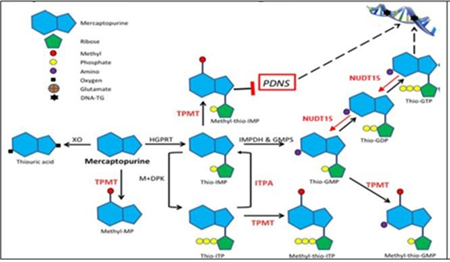
Figure 1 Metabolic pathways and mechanism of action of 6-Mercaptopurine.7
Pharmacogenomics 6-MP
The influence of the enzymes that participate in the metabolization of 6-MP has generated a branch of study aimed at pharmacogenomics and its application in the clinical setting. Single nucleotide polymorphisms (SNPs) of TPMT identifies that the prevalence is higher in the African and Caucasian, while in Chile frequencies of 8% in pediatric patients are described. Like those reported in countries of Caucasian and African origin.8 However according to the literature, the main SNP (rs116855232) in the NUDT15 gene is associated with a decrease in its activity, generating myelosuppression in patients who would ultimately be sensitive to the standard dose of thiopurines.9 This SNP presents a prevalence of 10% in the Asian origin,10 while some Latin American countries obtained prevalence of 8.8% in Uruguay and 12.5% in Mexico.9,11 The SNP rs3765534 was identified in the MRP4 gene which is reflected in the reduction of protein expression. Resulting in myelosuppression in patients treated with thiopurines. Allele frequencies of 18.7% are described in the Japanese population, 7.1% in the Chinese population and there are no data in Latin America.12 The SNPs are directly related to adverse events in both Asian and Latin American patients.13 Presenting intra- and inter-individual variability that affects the response to 6-MP which requires attention to analyze its pharmacogenomics. Both SNPs described above are not available as tests in clinical laboratories (LC) in Chile and the problem is that the available pharmacogenomic tests are insufficient requiring further development. Faced with this scenario the LC must be constantly updated and implementing methodologies to respond to the need for advances in medicine. In addition, new technologies are associated with greater efficiency, reduction of errors and better quality in the provision of services.14 The incorporation of molecular techniques such as polymerase chain reaction (PCR) has allowed an increase and better development of precision medicine. A term that appears more frequently in medical care demands15 being important in complex pathologies where being able to estimate the probability of a possible risk of presenting adverse events due to drugs has a real impact.16
The genetic variation in Chile is diverse and four ethnic groups are identified: native, African, European, and Asian, increasing the uncertainty in the response to chemotherapeutic drugs that we could find in our population.17 The individualization of the dose based on pharmacogenomics is essential to achieve the expected effects of the treatment being more effective and avoiding toxic effects. Therefore, implementing the detection of the NUDT15 and MRP4 SNPs by means of molecular biology techniques in LCs would allow the clinician to obtain more complete information on the response to drug treatment with 6-mercaptopurine.
Overall study design
Work based on an implementation and validation of molecular techniques carried out in the Clinical Laboratory of the Dr. Luis Calvo Mackenna Hospital (HLCM). The ethical regulations established by the HLCM were respected in addition to being approved by the Bioethics Committee for Research of the Faculty of Pharmacy of the University of Valparaíso.
Sample selection
71 blood samples (counter samples) collected in tubes with EDTA anticoagulant stored between 4° and 8°C were used. All samples were anonymized and recoded according to the institutional protocol for validation of HLCM clinical laboratory techniques.
Identification of genotypes
Automatic extraction of total nucleic acids from the commercial kit MagNA Pure Compact Nucleic Acid Isolation Kit I and the MagNA Pure Compact kit (Roche Diagnostics GmbH, Mannheim, Germany) were used according to the manufacturer's instructions. For both SNPs, the real-time PCR technique was used using two TaqMan® probes (Predesigned SNP Genotyping Assay for SNP NUDT15 and Drug Metabolism Enzyme Genotyping Assays for SNP gene MRP4), both probes were used together with TaqMan® Universal PCR. Master Mix (Applied Biosystems® - Termofisher), all the reagents were used according to the manufacturer's instructions and analyzed in the LightCycler® 480II Real-Time PCR System, (Roche) using the LightCycler 480 Software release 1.5.2.62 SP2.
Validation of techniques
Five analyzed and resolved samples were used in this study; three for the NUDT15 gene (native, heterozygous, and mutated homozygous), by means of the primers F: 5´-CCCCTGGACCAGCTTTCTG- 3´ and R:5´-CCACCAGATGGTTCAGATCTTCTTTAAA-3´ (IDT, USA, Fermelo) and two for the gene MRP4 native and homozygous for the variant) by means of the primers F: 5´-TCCAGTGGCTGATTTTCTGA- 3´ and R: 5´-GAGTGTAAACTGCGGTGGT-3 (IDT, USA, Fermelo).
Conventional PCR for both techniques was performed with GoTaq Green Master Mix (Promega, USA) separated by 2% agarose gel electrophoresis, using a Bio-Rad Power Pac Basic DNA electrophoresis chamber (California, USA) together with a molecular size AccuRuller 1KB DNA Ladder (GeneBio Systems, Canada) and were visualized after staining with Gelred 1000x Nucleic Acid Stain (Biotium, USA) using a TCP-26.LMX UV transilluminator (Vilber Lourmat, France).
The samples were sequenced by the Sequencing Service of the Faculty of Biological Sciences of the Pontificia Universidad Católica de Chile. The sequences obtained were contrasted with reference sequences from the National Center for Biotechnology Information (NCBI) using the Chromas DNA Sequencing program (Technelysium, Australia) and BioEdit 7.2. With the results obtained, the genotypic frequencies for both polymorphisms, the allele frequencies, and the Hardy-Weinberg (H-W) equilibrium were calculated.
For the SNP rs116855232 of NUDT15, the three possible genotypes were found. The size of the amplicon (450bp) was verified and once the three samples were sequenced. The results obtained were compared with the reference gene according to the NCBI database. Its result, proving that 99% homology is achieved, due to the substitution of T for C. The alignment of the native gene amplified and sequenced in this study presents 100% homology with its counterpart in the database NCBI (Table 1).
|
Native gen |
Heterozygous gene |
Homozygous gene |
Real-time PCR |
C/C |
C/T |
T/T |
Sequencing |
C/C |
C/T |
T/T |
Table 1 Comparison of results of the real-time PCR analysis and sequencing for NUDT15
Samples analyzed by real-time PCR using TaqMan® probes compared to sequencing method.
For the SNP rs3765534 of MRP4 two homozygous genotypes were found. For the native gene and the variant studied. The size of the amplicon was consistent with what was theoretically expected (600 bp). It was compared with the reference gene according to the NCBI database. The alignment of the native gene amplified and sequenced in this study presents 100% homology with its counterpart in the NCBI database (Table 2).
|
Native gen |
Homozygous gene |
Real-time PCR |
G/G |
A/A |
Sequencing |
G/G |
A/A |
Table 2 Comparison of results by real-time PCR analysis and sequencing for MRP4
Samples analyzed by real time PCR using TaqMan® probes compared to sequencing method.
The genotypic frequencies for both polymorphisms were 12.6% heterozygous in NUDT15 rs116855232 C/T. While the homozygous T/T genotype was found in 1.4% of the population analyzed. For MRP4, the rs3765534 G/A variant was detected in a single sample corresponding to 1.4% of the total analyzed. Allele frequencies were 7.7% for the rs116855232 (T) allelic variant in NUDT15 and 1.4% for the rs3765534 (A) allelic variant in MRP4. Regarding the Hardy-Weinberg (H- W) equilibrium. The SNP rs116855232 NUDT15 is in equilibrium (p=0.07), while rs3765534 MRP4 is not (0.00) (Table 3).
Samples analyzed n=71 |
|||||||
|
Allele frequency |
Genotype frequency |
H-W balance |
||||
SNPs |
Reference allele |
Variant allele |
Homo. Native |
Heterozygous |
Homo. Variant |
X2 |
p |
NUDT15 |
C=0,92 |
T=0,08 |
C/C=0,86 |
C/T=0,13 |
T/T=0,01 |
3,13 |
0,07 |
MRP4 |
G=0,99 |
A=0,01 |
G/G=0,99 |
G/A=0,00 |
A/A=0,01 |
15,14 |
0,00 |
Table 3 Frequency and Hardy-Weinberg for SNP NUDT15 415C>T and MRP4 2269G>A
None.
Authors declare that there is no conflict of interest.

©2022 Morales, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.