eISSN: 2574-9927


Research Article Volume 7 Issue 4
1Department of Physics, Federal University of Health Sciences Azare, Nigeria
2Department of Physics, Federal University of Technology Minna, Nigeria
3Department of physics with electronics, Federal University Birnin Kebbi, Nigeria
Correspondence: Samaila B, Department of physics with electronics, Federal University, Birnin Kebbi, Nigeria
Received: November 20, 2023 | Published: December 11, 2023
Citation: Ahmed H, Michael DO, Samaila B. Current challenges of the state-of-the-art of AI techniques for diagnosing brain tumor. Material Sci & Eng. 2023;7(4):196-208. DOI: 10.15406/mseij.2023.07.00224
Background: Brain is the control center of the human body, in recent time, different variety of brain diseases are being discovered. The brain disease diagnosis tools are becoming challenging and still an open area of research, application of AI in brain disease diagnosis has made disease prediction and detection more precise and accurate. Automated technologies for non-invasive analysis of brain images have become necessary, because disease of brain is fatal and are the cause of large number of deaths in developed countries. Brain tumor surgery augmented with AI can result in safer and more effective treatment. The knowledge gap between clinical and data science experts still presents significant challenges. This paper will review literatures related to current challenges of AI technologies for brain tumor diagnosis and suggest new directions of AI technologies for diagnosing brain tumour. A systematic search of major academic databases (such as Science Direct, IEEE explore digital Library, and Google scholar) was conducted to identify relevant studies published between 2015 and 2023. The search term used in this study include “Brain tumor Diagnosis”, “AI challenges in Brain tumor Diagnosis”, ‘AI techniques” and “AI challenges in medicine and future”. Studies were included if they utilized AI techniques for brain tumor diagnosis. The identified studies were evaluated for the key challenges they encountered in their diagnostic approaches. The Present study identified several challenges related to the application of AI techniques in brain tumour diagnosis. These challenges include: Interpretability and explainability, variations in tumour location, shape, and size which make accurate segmentation and classification difficult. Overall, the challenges in explaining brain tumor detection stem from the unique requirements and complexities of the healthcare domain, necessitating specialized techniques and approaches. This study summarizes the new directions for AI as (I) Data Hungry: Large, standardized, annotated data sets and excellent ground truth data are necessary for the development of accurate AI. (II) Radiomics: makes it possible to extract a vast number of quantitative features from intricate clinical imaging arrays and convert them into high-dimensional data that can be further processed to determine their relationship to the histological features of the tumor, which represent underlying genetic mutations and malignancy as well as grade, progression, response to therapy, and even overall survival (OS). (III) Black box: AI, for instance, is capable of predicting the best course of care for a patient, but it is unable to explain its reasoning. A trend toward easing this restriction is interpretable deep learning, (IV) Demonstrating the generalizability of deep learning applications and conducting external validation are two major obstacles. (V) There are knowledge gaps in clinical oncology that need to be filled in order to successfully integrate AI and maximize its effects. (VI) Several national professional bodies have started programs to bridge these knowledge gaps and advance the adoption of AI in oncology in response to these difficulties.
Keywords: artificial intelligence, brain tumor, diagnosis
The industrial revolution 4.0 and technological advancement are in wide spread application across all discipline, information and communication technology such as artificial intelligence (AI), internet of thing (IOT) and block chain technology are accelerating and solving complex problems in Health care system.1 stated that doctors can stay current on patient data, offer virtual help, and send emergency answers when necessary thanks to block chain and AI.2 The problems of record keeping, ongoing patient monitoring, long-distance patient care, and emergency response are all solved by IoT devices. Furthermore, the transparency of blockchain technology can strengthen clinical trial data integrity and boost confidence in research findings. In conclusion, applications of block chain and artificial intelligence (AI) in the delivery of healthcare include medical supply chain management, drug development and clinical trials, telemedicine and remote patient monitoring, precision medicine and genomic data, secure and interoperable health records, healthcare payment and insurance claims, and so on. Obstacles & Things to Think About are: Interoperability Standards, Regulatory Compliance, Data Privacy and Consent, cost, technology adoption and reliability and maintainability.
AI is the study of how to use computer to mimic human intelligent behavior, such as learning, judgement and decision making through training using large amount of data.3 The advancement of AI technologies helps clinical experts to facilitate more efficient and effective electronic healthcare systems to the patients.4,5 The formation of abnormal cells in or near the brain lead to the start of brain tumor and consequently affect patient healthcare.6,7 Automated technologies for non-invasive analysis of brain images have become necessary, because disease of brain is fatal and are the cause of large number of deaths in developed countries. Both adults and children are included in the American Cancer Society's predictions for brain and spinal cord cancers in the country for 2022. There will be a total of 25,050 malignant brain or spinal cord tumor diagnoses (14,170 for men and 10,880 for women). If benign tumors (tumors other than cancer) were included, these figures would be substantially higher. An estimated 18,280 individuals (10,710 men and 7,570 women) will pass away due to brain and spinal cord malignancies.8 Despite the significant developments in molecular biology and brain tumor imaging, including MRI, CT, PET, DTI, and SPECT. There are currently no easily accessible automated systems for brain imaging diagnosis in clinical practice.9 The brain is the human body's control center, and a wide range of brain disorders are being identified these days. Furthermore, there is still much to learn about the diagnostic techniques for brain diseases, which is growing more difficult. The use of AI in brain disease diagnosis has improved the accuracy and precision of disease detection and prediction.10
Theoretical background of the AI
Human-machine intelligence (AI) One of the current uses of artificial intelligence (AI) is machine learning (ML), a branch of computer science that focuses on building intelligent machines that mimic human behavior. ML is based on the notion that we should give machines access to data so they can learn on their own. DL is a subfield of ML. While deep learning is more akin to animal vision, machine learning is more like human vision. Convolutional Neural Network (CNN) is a new method for image analysis using deep learning. Computer assisted diagnostic (CAD) technologies process digital pictures to highlight certain noticeable disorders to help radiologists or other medical practitioners.11 As displayed in Figure 1.
Figure 2 displays the five major subdomains of artificial intelligence. There are several possible clinical uses for each subdomain of AI in brain tumor surgery. There are many more subfields within AI, hence this diagram is not all-inclusive. In12 the dependent variable (the target) and the independent variable (a collection of predictors) make up supervised learning. These variables are used to create a function that maps inputs to intended outputs. The model is trained repeatedly until it reaches a high degree of accuracy on the training set. kNN, logistic regression, decision trees, random forests, regression, and so on are a few instances of supervised learning. There won't be any target variable to forecast in unsupervised learning. Examples of unsupervised learning include k-means, apriori algorithms, and others. The machine is placed in an environment where it continuously educates itself through trial and error in reinforcement learning. In this case, the computer attempts to get the knowledge necessary to make accurate decisions by learning from its prior experiences. Markov decision process serves as one illustration of reinforcement learning (Figure 3).13

Figure 2 The five key subdomains of Artificial intelligence.12
Deep learning has medical research areas as shown in Figure 4.
The evolution of ML and DL in the healthcare industry for brain disease diagnosis and detection has many approaches over the time as shown in Figure 5. The distinction between ML and DL from the literature can be seen in the information recognition pattern15 as shown in Figure 6.
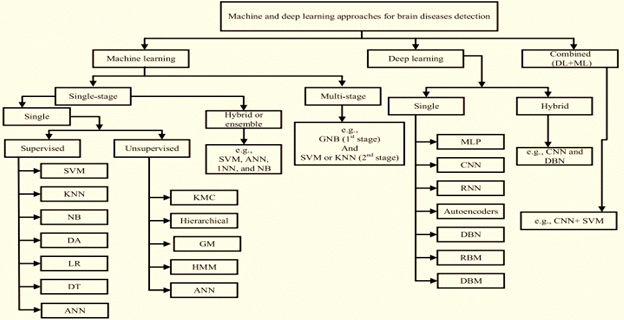
Figure 5 Classifications of ML and DL techniques to detect brain diseases.14
Background knowledge of brain tumor
Brain is the central processing and control center in human body.14 The formation of abnormal cells in the brain or near the brain lead to the start of tumor called brain cancer, the abnormal cells altered the brain normal brain processing ability and consequently affects the patient’s health.16 Brain tumor as masses of abnormal cells (tissues) growing out of control can be classified according to starting locations, adverse effect, growing level and starting cells Primary tumors start in the brain and secondary (metastatic) tumor started in somewhere in the body and reach out to brain.17 Benign (non-cancerous) tumors do not grow into nearby tissues or distant tissues, while malignant (cancerous) tumors can spread into nearby tissues or distant tissues. Both benign and malignant brain tumors can spread through brain tissue, but they rarely spread to other parts of the body. While brain tumors that grow slowly, such as Grade I and Grade II, rarely invade nearby tissues, brain tumors that grow quickly, such as Grade III and Grade IV, mostly can.18 The brain tumor types based on the starting cells are shown in Table 1.
|
Brain tumor |
Starting cell |
Prevalence |
|
Glioma |
Glial cells |
The most prevalent type of glial cell-derived central nervous system (CNS) tumor is called a glioma. Gliomas are detected in six cases per 100,000 persons in the United States each year. |
|
Meningiomas |
Meninges cells |
They occur more frequently in women and older persons, developing in about 8 out of every 100,000 people annually. |
|
Medulloblastoma |
Neuroectodermal cells |
About 20% of all pediatric brain tumors and 63% of intracranial embryonic tumors are caused by medulloblastoma. These tumors have an overall yearly frequency of about 5 cases per million in the pediatric population. They can develop from childhood and into adulthood. |
|
Ganglioglimas |
Both neuron and glial cells |
Rare combined glio-neural tumors called gangliogliomas (GGs) account for 0.4% of central nervous system neoplasms and 1.3% of all primary brain tumors. |
|
Schwannomas (neurilemmomas) |
Schwann cells |
The incidence is 4.4 to 5.23 cases per 100,000 adults/year; in children and adolescents, it is 0.44 cases per 100,000/year. |
|
Craniopharyngiomas |
Pituitary gland |
In the United States, an estimated 350 new cases of craniopharyngioma are diagnosed each year, resulting in an age-adjusted incidence of 0.19 per 100,000 persons |
Table 1 The brain tumor types based on the starting cells
Brain tumor diagnostic imaging modalities
Magnetic resonance imaging (MRI)
Imaging with magnetic resonance (MRI) is a non-invasive method that uses magnetic field to generate radiofrequency (RF) field to produced images of soft tissue with high resolution. It was described in 1930s and 40s. the principle of MRI is based on the interactions of protons (hydrogen atoms), strong magnetic fields and radiofrequencies of different energies. (Figure 7).19
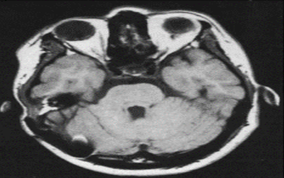
Figure 7 MRI brain scan.20
Patients is placed inside the strong magnet at still position to reduce motion artefacts in the image. MRI produced detailed anatomical structure of the brain,20 spinal cord and other body parts, it has the advantage of being able to visualized anatomical images in three planes: axial, sagittal and coronal views.21 MRI has the advantages of giving higher soft tissue contrast and being able to detect blood flows and cryptic vascular malfunctions, MRI has the advantage of being free from ionizing radiation exposure over computed tomography (CT) and conventional x-ray machines. T1-weighted, T2-weighted, diffusion weighted imaging (DWI), Proton density (PD-weighted), Fluid attenuated inversion recovery (FLAIR), Auto-calibrating reconstruction for cartesian imaging (ARC), and Generalized Auto-calibrating partial parallel acquisition (GRAPPA) are the imaging sequences used in magnetic resonance imaging (MRI). ARC is multi-coil parallel imaging (PI). Based on the time to echo (TE) and repetition time (TR), T1-weighted and T2-weighted are generated. T1-weighted has longer TE and TR. Another difference is by looking at cerebrospinal fluid (CSF), CSF is darker in T1-weighted and bright in T2-weighted images. The FLAIR sequence is mainly T2-weighted image with longer TE and TR. Diffusion weighted image is used to detect random movement of water proton. Proton density is in between T1 and T2 with pulse sequence of long TR and short TE.21 MRI is broadly classified into structural magnetic resonance imaging (sMRI) and functional magnetic resonance imaging (fMRI), sMRI is mostly applicable to clinical practices and research purposes. The distinction between sMRI and fMRI is difficult to make as function and structure are closely related, from biological view point fMRI provides dynamic physiological information which includes blood oxygen level depended (BOLD), perfusion and blood flow, while, sMRI displays static anatomical information which include studies of epilepsy, schizophrenia, dementia, trauma, tumours and multiple sclerosis.22 Structural MRI sequence has high contrast between gray matter and white matter giving room for volume quantification of the gray and white matter. The common method used for processing sMRI is voxel-based morphometry, it can also be used to assess the degree of cortical folding or pattern and variation of cortical gyrification. sMRI has the advantages of clear interpretation, early implemented across centres and relatively low cost over fMRI, electroencephalography and proton magnetic resonance spectroscopy23 (Figure 8).
Computerized tomography (CT)
The brain's structure, including details like blood perfusion, can be seen by computerized tomography scans using X-rays. The resulting images are two-dimensional and have a very low resolution, although since 1998, the quality has significantly increased. Better technology has allowed for the creation of multisections and eight times faster speeds, resulting in well-defined three-dimensional images from a single section. Underdeveloped brain regions or locations of impact, tumor, lesion, or infection may be visible on a CT scan.24
Positron emission tomography (PET)
Using positron emission tomography scanning, one can obtain a three-dimensional picture of the brain's functional activities in addition to its structural makeup. Fluorodeoxyglucose, a radioactive sugar tracer, must be subcutaneously injected into the patient's bloodstream in order for PET imaging to be performed. Gamma-rays, a type of electromagnetic radiation with a higher energy than X-rays, are produced by radioactive material. The radioactive substance enters the brain and travels throughout the body. Pairs of gamma rays are indirectly emitted by the positron-emitting radionuclide (tracer) in each area of the brain being examined, and they are detected using a ring of detectors outside the head.4,25
Single photon emission computed tomography (SPECT)
In single photon emission computed tomography, two or more synchronized gamma cameras record the signals from gamma rays (rather than when the emissions are opposite at 1800). Multiple 2-D pictures are generated and tomographically rebuilt to 3-D. While a segment can be viewed from multiple perspectives, it is not as clear as a PET image. Using more readily available, longer-lived radioisotopes, SPECT scanners are less costly than PET scanners. The brain's blood flow may be traced to determine the areas of metabolic activity, which facilitates the evaluation of brain functions.26
Diffusion tensor imaging (DTI)
A kind of diffusion magnetic resonance imaging (MRI) called diffusion tensor imaging is used to watch brain activity as it happens. A common method for imaging white matter in the brain is to detect the limited diffusion of water through the tissue under study (Table 2).27
|
Reference |
Year |
Datasets |
AI technology |
|
2021 |
Commonly used brain tumor dataset |
Deep neural network and residual network |
|
|
2021 |
BRASTS 2013 Challenges |
CNN |
|
|
2018 |
BRASTS 2012, ISLES 2015 |
DNN |
|
|
2018 |
BRASTS 2021 |
Orthogonal gamma distillation machine learning model |
|
|
2020 |
MICCAI datasets and BRASTS 2015-2017 |
Grab cut methods |
|
|
2020 |
MRI data |
CNN-based and AlexNet |
|
|
2021 |
MRI T1-W and T2-W |
Auto ML |
|
|
2020 |
MRI BRASTS |
Adaptive KNN |
|
|
2020 |
MRI BRASTS |
RF |
|
|
2020 |
MRI BRASTS |
SVM |
|
|
2020 |
MRI GBM |
CNN+SVM |
|
|
2020 |
MRI BRASTS and ISLES |
LSTM+Softmax |
|
|
2021 |
MRI BRATS 2018 |
3D-CNN |
|
|
2020 |
MRI Figshare |
Inception V3 softmax DenseNet+softmax |
|
|
2020 |
MRI T1 Figshare |
Hybride CNN-NADE |
|
|
2020 |
MRI T1 |
CNN-ELM |
|
|
2020 |
MRI (TCGA-GBM) |
SequenceNet CNN-Elm |
|
|
2020 |
MRI Kaggle Repository |
CNN |
|
|
2020 |
MRI Kaggle |
BranMRNet CNN |
|
|
2020 |
MRI BRTASTS |
Stacked sparse auto encoder+softmax |
|
|
2020 |
MRI BRASTA |
Deep CNN |
|
|
2021 |
MRI T1, T2 and Flair |
DBM |
|
|
2020 |
MRI BRASTS |
CNN-VUG19+KNN CNN-VUG19+Ensemble |
|
|
2021 |
MRI BRASTS |
3D CNN |
|
|
2019 |
TCGA-GBM |
NS-CNN NS-EMFSE |
|
|
2020 |
MRI BRASTS |
ELM |
|
|
2018 |
MRI BRASTS |
Two-pathway group CNN |
|
|
2019 |
MRI |
KNN, ANN and SVM |
|
|
2021 |
Figshare |
denseNet-41-b with cornerNet |
|
|
2019 |
MRI T1-W |
RT |
|
|
2019 |
76 MRI |
SVM |
|
|
2019 |
126-MRI |
RF |
|
|
2019 |
MRI and PET |
SVM |
|
|
2019 |
233-MRI |
CNN |
|
|
2020 |
500-MRI |
CNN |
|
|
2019 |
100-MRI |
RF-SVM |
|
|
2019 |
180-MRI |
DNN |
|
|
2019 |
350-PETs |
Deep belief network |
|
|
2018 |
32-MRI |
Fuzzy C-means |
|
|
2018 |
30-MRI |
RF |
|
|
2018 |
9-MRI |
SVM |
|
|
2019 |
64-MRI |
CNN |
|
|
2019 |
60-MRI |
Supervised learning LOCATE |
|
|
2015 |
10-pateints MRI |
Hybrid level set |
|
|
2015 |
BTASTS |
Fully automated generic method |
|
|
2015 |
SPES and SISS |
EM |
|
|
2017 |
BRASTS |
Otsu algorithm |
|
|
2017 |
21HGG patients |
Non-negative matrix factorization |
|
|
2019 |
1340-clinical MRI |
Adaptive thresholding |
|
|
2021 |
Local Data |
SVM, CNN |
Table 2 Summary of the related literature
Diffuse optical tomography (DOT)
A non-invasive imaging method called diffuse optical tomography (DOT) uses near-infrared light to scan the inside of the brain for changes in oxygenation and other physiological parameters that may have resulted from a stroke, seizure, or hemorrhage. Despite having a lower spatial resolution than MRI, DOT has the advantage of being simpler and faster at taking measurements. The devices are small and lightweight, roughly the size of a laptop and a small suitcase, making them easy to carry to the patient's bedside for ongoing brain activity monitoring.28
The period considered in study for the literature review is from 2015 to 2023, the databases used to obtain the literatures are Science Direct, IEEE explore digital Library, and Google scholar. The search criterions are “Brain tumor Diagnosis”, “AI challenges in Brain tumor Diagnosis”, ‘AI techniques” and “AI challenges in medicine and future”. The inclusion criteria (IC) and exclusion criteria (EC) are shown in table below (Tables 3,4).
|
Inclusion criteria |
Eexclusion criteria |
|
|
|
|
IC1: Paper must from 2015 to 2022 and peer reviewed |
EC1: duplicate studies in different database |
|
IC2: paper should use only MRI, CT or PET for Imaging acquisition |
EC2: case study papers |
|
IC3: paper should have automated AI technology |
EC3: Study less cited by the peer reviewed papers |
|
|
EC4: Study using imaging other than MRI, Ct or PET |
Table 3 Selection criteria
Challenges in explaining brain tumor detection
Brain tumor detection poses several challenges in terms of explanation. Existing explanation techniques for image classifiers, such as ImageNet, may not be adequate for explaining the detection of brain tumors in MRI brain images.76 The variations in tumor location, shape, and size make accurate segmentation and classification difficult.77 Additionally, the complexity of the brain as an organ and the critical nature of brain tumors contribute to the challenge of early detection and diagnosis.78 Further improvement is required in the efficiency of existing detection schemes, and critical research challenges need to be addressed in order to develop new methods for brain tumor detection.79 Overall, the challenges in explaining brain tumor detection stem from the unique requirements and complexities of the healthcare domain, necessitating specialized techniques and approaches. The issues surrounding the diagnosis and assessment of brain tumors have been discussed by,80 The random forest classifier approach finds the tumor in less machine time and with measured precision in their suggested treatment model. Our research revealed that the suggested system has a high accuracy rate for detecting tumors, a high rate of diagnosing diseases, and a low computing time for detecting diseases.
Kenneth Aldape 81 reported that, in an effort to promote advancements in the knowledge and capacity to effectively treat brain tumor patients, Cancer Research UK assembled a global panel of physicians and scientists working in laboratories to pinpoint obstacles that need to be surmounted in order to cure every patient with a brain tumor. The seven main issues are outlined here to provide future research and funding priorities. These include: (1) revamping the pipeline for brain tumor research and therapy; (2) utilizing the entire field of neuroscience; (3) comprehending the function of the microenvironment in the physiology and treatment of brain tumors; and (4) creating more accurate preclinical models. (5) find drugs for challenging targets in a diverse environment (6) create a precision medicine strategy for treating brain tumors (7) reduce treatment for Less-aggressive brain tumours.
Kenneth Aldape79 employed a Weiner filter with several wavelet bands to improve and de-noise the input slices, Using Potential Field (PF) clustering, subsets of tumor pixels are identified. Furthermore, in Fluid Attenuated Inversion Recovery (Flair) and T2 MRI, the tumor zone is isolated using a global threshold and several mathematical morphological techniques. Features from the Gabor Wavelet Transform (GWT) and Local Binary Pattern (LBP) are combined for reliable classification. Outcomes Peak signal to noise ratio (PSNR), mean squared error (MSE), and structural similarity index (SSIM) are used to evaluate the suggested technique. The findings are as follows: 76.38, 0.037, and 0.98 on T2 and 76.2, 0.039, and 0.98 on Flair, respectively. Pixels, individual features, and fused features have all been utilized to evaluate the segmentation outcomes. The suggested method is compared at the pixel level to ground truth slices and validated in terms of error region (ER), pixel quality (Q), background (BG), and foreground (FG) pixels. Using a local dataset, the method produced precision values of 0.93 FG, 0.98 BG, and 0.010 ER. BRATS 2013, a multimodal brain tumor segmentation challenge dataset, yields precision values of 0.93 FG, 0.99 BG, and 0.005 ER. Similarly, 0.015 ER, 0.97 FG, and 0.98 BG accuracy are obtained on BRATS 2015. The average Q value and variance in terms of quality are 0.88 and 0.017, respectively. Particularity, sensitivity, accuracy, area under the curve (AUC), and dice similarity coefficient (DSC) at the fused feature-based level are, respectively, 1.00, 0.92, 0.93, 0.96, and 0.96 on BRATS 2013, 90, 1.00, 0.97, 0.98, and 0.98 on BRATS 2015, and 90, 0.91, 0.90, 0.77, and 0.95 on local dataset. The suggested method performed better than the current methods.
New directions of AI technology for brain tumor diagnosis
Brain tumors are incurable diseases that impact nerves and human blood cells due to aberrant brain cell development. As it can help physicians plan surgeries, early and accurate brain tumor diagnosis is crucial to avoiding difficult and unpleasant treatment procedures.82 AI-enhanced brain tumor surgery can lead to safer and more efficient care.83 There are still many difficulties due to the knowledge gap that exists between data science and healthcare specialists. In contrast to data scientists, who possess advanced cognitive skills in data science to comprehend AI mechanisms, physicians have extensive experience in oncologic workup and management. To close the gap between clinical and data science professionals, more collaboration should be encouraged. The role of AI is another crucial matter. Without expertise, it is nearly impossible to run an AI. AI shouldn't be used in an entirely unsupervised setting as a stand-alone solution. Conversely, it is a useful tool that can assist in areas where human talents are still limited and a beneficial assistance to experts.83
Radiomics
Radiomics makes it possible to extract a vast number of quantitative features from intricate clinical imaging arrays and convert them into high-dimensional data that can be further processed to determine their relationship to the histological features of the tumor, which represent underlying genetic mutations and malignancy as well as grade, progression, response to therapy, and even overall survival (OS). In contrast to conventional brain imaging, radiomics offers quantifiable data associated with significant biologic features and the use of deep learning, which illuminates the complete automation of imaging diagnosis. Recent research has demonstrated the wide range of applications of radiomics, including the identification of primary tumors, differential diagnosis, grading, assessment of aggression and mutation status, and prediction of treatment response and recurrence in brain metastases, pituitary tumors, and gliomas.84
Radiomics is a rapidly expanding field and is still in extensive clinical exploration stage, with many obstacles to overcome. Current standards lack results validation, incomplete results reports, and unidentified confounding variables in the source database, especially for retrospective data.85,86 radiomics and radiogenomics can only identify the correlation, thus lacking robustness and credibility without tissue biopsy.87 The auto segmentation procedures used today are dispersed and lack standardized practices. According to the research we looked at, brain tumor radiomics rarely uses more sophisticated algorithms, like deep learning, than lung, prostate, or colorectal cancer radiomics.88 Current research in brain tumor still lack huge populations, especially from several sites.89 Furthermore, there are ethical concerns regarding the motivation of academics and governments to share personally validated data for machine learning, even when the creation of AI algorithms necessitates not just basic technology but also legislation and maybe ethics.85
Data hungry
One continuous need for AI is the collection of a sizable, publicly available, well-annotated cancer dataset. Good data is essential for the effective creation of an AI model. Even if there is a rising volume and variety of data available, the evaluation of data quality is not standardized. AI is challenging to use since patients have a wide range of cancer types and frequently lack clinical, imaging, or genetic data. AI is challenging to use since patients have a wide range of cancer types and frequently lack clinical, imaging, or genetic data. Equity and Access to Data The issues of overfitting are directly caused by restrictions on the availability and caliber of data. More than any other ML approach, DL neural networks need a lot of data. This can be problematic for the healthcare industry when trying to apply AI to less common disease processes. Moreover, data silos can exist inside certain institutions. Concerns about the transmission of protected patient health information, the absence of an infrastructure for data sharing between institutions, the variability and incompleteness of data collecting, and competition between institutions are all factors contributing to this relative data drought. With an increasing focus on expedited data collecting90 and several multi-institutional data-sharing agreements,91,92 these challenges are starting to be addressed. Research organizations can now publish their own data, which may encourage openness.94 Guidelines for FAIR (findable, accessible, interoperable, and reusable) data utilization have also been presented.93
Large, standardized, annotated data sets and excellent ground truth data are necessary for the development of accurate AI. The majority of clinical studies for gliomas are multi-institutional, which makes it more difficult to get consistent data sets,27,95 Understanding what sorts of datasets are required for a possible utility and how to get these datasets is crucial to optimizing the intended results. Commonly, radiographic imaging, cancer genome, medical records, pharmacological information, and biomedical literature are among the sensitive and helpful indications or features for AI-powered cancer research.96,97
Black box
The model's relative opacity limits the application of AI in practical situations. The machine was unable to explain how or why it had come to this conclusion. The "black box" dilemma is how people frequently refer to this.98 It is challenging to identify the input data features that contribute to the result. AI, for instance, is capable of predicting the best course of care for a patient, but it is unable to explain its reasoning. A trend toward easing this restriction is interpretable deep learning.99,100 The mystery box issue Although these models consistently achieve good performance, one of the main obstacles to the use of AI in healthcare is the worry that they are relatively opaque. For example, based on a patient's prior two years of EHR data, a DL model may correctly predict that the patient will acquire pancreatic cancer. But why did the model make that prediction? We can currently only deduce a limited amount of the exact reasoning underlying DL-based predictions. This issue is frequently called the "black box" dilemma.101 Understanding the reasoning behind each clinical choice has long been crucial in the practice of medicine. Conventional machine learning methods, such as linear regression, are not very good at modeling intricate relationships, but they are straightforward to understand since they provide us with a collection of pre-defined features and the feature weights that represent their respective impact sizes. On the other hand, unstructured input data is used in deep learning, and the majority of knowledge creation takes place in the hidden layers. As a result, identifying the precise attribute or characteristics of the input data that influenced the result becomes challenging. The use of AI-based algorithms in healthcare will be significantly impacted by this interpretability issue, from both a practitioner and a regulatory standpoint.102–105
Despite the fact that AI algorithms employ a wide range of features in their decision-making, many of the previously published AI algorithms cannot be easily replicated by other researchers due to the complexity of their analytical methods. The "blackbox" aspect of AI is being investigated further, and the results could one day make it possible to follow otherwise opaque processes step-by-step using transparent methods.106,107 Currently, addressing the black box issue is a key area of research attention.3 Many techniques have been developed for AI image analysis algorithms, such as saliency maps, class activation mapping, feature visualization, and sensitivity analyses, in which specific areas of the image are hidden to reduce prediction error.108 Even though these techniques have improved in recent years, more research is required to fully understand the reasoning behind deep neural network decision-making.
Proving generalizability and real-world applications
Even though artificial intelligence (AI) is being quickly used to oncologic research, more has to be done to convert these findings into practical, therapeutically useful applications. Demonstrating the generalizability of deep learning applications and conducting external validation are two major obstacles. Neural networks have a strong propensity to produce overfitted models that do not generalize across various populations because to their complexity and astronomically high parameter counts (sometimes in the millions). Furthermore, several external validation sets would be needed to demonstrate the effectiveness of an application due to the notable variety of medical data among institutions.109 One of the main obstacles inhibiting the AI algorithms' wider clinical application is their generalizability. The majority of AI applications used in gliomas and oncology to yet have been trained on patient populations that are still quite small. The huge and diverse population of gliomas makes the performance of an AI algorithm created on a small population suboptimal.110
Education and expertise
There are knowledge gaps in clinical oncology that need to be filled in order to successfully integrate AI and maximize its effects. Physicians are now undertrained in data science and machine learning, which hinders their capacity to comprehend deep learning mechanisms, choose suitable algorithms, and carry out research.111 Analogously, the majority of data scientists lack expertise in oncologic workup and management, which hinders their capacity to recognize significant and appropriate clinical use cases. Clinical oncologic departments and bioinformatics and data science divisions should work together more, and when necessary, strategic alliances with technological companies should be established. The connection between clinicians and engineers presents another barrier to the widespread adoption of AI in gliomas and cancer. Presently, the majority of computer scientists are unfamiliar with the complexities of clinical patient management, while physicians have relatively little training in computer/data science.112
Promoting AI in oncology: professional societies and national initiatives
Several national professional bodies have started programs to bridge these knowledge gaps and advance the adoption of AI in oncology in response to these difficulties. The American College of Radiology (ACR) established the ACR-DSI, or American College of Radiology Data Science Institute, to work with industry, government, and radiologists to advance AI in imaging.113 The ACR-DSI encompasses multiple fundamental objectives: (i) offering guidelines for gauging AI algorithm performance ("Touch-AI"), (ii) autonomous, external verification of algorithms and managing the regulatory environment ("Certify-AI"), and (Assess-AI"), a long-term, prospective assessment of implemented algorithm performance. In addition, a number of use cases for suggested AI imaging applications with unmet clinical needs have been established by the ACR-DSI. In collaboration with oncologists, industry, and academia, the American Society of Clinical Oncology (ASCO) and American Society for Radiation Oncology (ASTRO) have launched a big data initiative called Cancer Link. The initiative aims to provide oncologists with user-friendly knowledge dissemination while tracking and evaluating treatment outcomes in real-time.114 The foundation of the project is an ever-expanding database of de-identified patient data that can be searched through and examined. ASTRO and Cancer Link teamed in 2017 to offer radiation oncology knowledge and database applications. Additionally, one of the main goals of the ASTRO Research Agenda for 2018 is bioinformatics and big data analytics.115 National Institutes of Health (NIH): To encourage the development of tools for integrating big data and data science into biomedical research, the Big Data to Knowledge (BD2K) effort was started as part of the NIH Common Fund.116 Using pre-existing national datasets, such as The Cancer Genome Atlas (TCGA) and the Library of Integrated Network-based Cellular Signatures (LINCS), and machine learning (ML) techniques to find patterns in the data that could lead to previously unidentified compounds for cancer therapeutics is one of the initiative's main focuses (Tables 5,6).3
|
Brain tumor association |
Website |
|
National brain tumor society |
https://braintumor.org/ |
|
The American Brain Tumor Association |
https://www.abta.org/ |
|
The International Brain Tumour Alliance (IBTA) |
https://www.cancer.gov/rare-brain-spine-tumor/living/related-organizations |
|
Voices Against Brain Cancer's (VABC) |
http://www.voicesagainstbraincancer.org/ |
|
European Organisation for Research and Treatment of Cancer EORTC |
https://www.eortc.org/research_field/brain/ |
Table 5 Brain tumor associations around the world
|
Brain tumor database |
Website |
|
Figshare Brain datasets |
https://figshare.com/articles/dataset/brain_tumor_dataset/1512427 |
|
BRASTS 2012-2018 Challenge datasets |
https://paperswithcode.com/dataset/brats-2018-1 |
|
Kaggle datasets |
https://www.kaggle.com/search |
|
Brain tumor datasets IEEE |
https://ieee-dataport.org/documents/brain-tumor-dataset |
|
The Cancer Genom Atlas (TCGA) |
https://portal.gdc.cancer.gov/projects/TCGA-GBM |
|
The Cancer Imaging Archive (TCIA) |
https://www.cancerimagingarchive.net/ |
|
Internet Brain Segmentation Repository (IBSR) |
http://allie.dbcls.jp/pair/IBSR;Internet+Brain+Segmentation+Repository.html
|
|
Brain Web Simulated Datasets |
http://allie.dbcls.jp/pair/IBSR;Internet+Brain+Segmentation+Repository.html |
|
Ischemic Stroke Lesion Segmentation Challenge 2015-2017 Datasets |
http://www.isles-challenge.org/ISLES2015/ |
|
Harvard Medical School Whole Brain datasets |
https://www.med.harvard.edu/aanlib/ |
Table 6 Brain tumor database around the world
Each of the brain tumor database have their specific ethics related to data sharing for machine learning which the researcher needs to explore for compliance.117
Obstacles in the treatment of primary brain tumors at various tumor growth phases
The ventricular–subventricular zone's non-malignant cellular makeup contains neural stem cells, which proliferate and give rise to transit-amplifying cells. Neuroblasts that migrate are derived from transit-amplifying cells90. The neural stem cell niche also contains ependymal cells. The niche may interact with other cell types, such as microglia and astrocytes, and is closely linked to blood arteries. When the non-malignant hierarchy starts to change, the premalignant proliferation of transit-amplifying cells and migratory neuroblasts is probably caused by the malignant transformation of neural stem cells. This disordered hierarchy is what causes the cancerous brain tumor. The ultimate goal of treating these lesions is to eliminate the tumor cells in order to bring about a cure. The primary research topics at each stage, from the genesis of cancer to its remission after successful therapy, are indicated by the blue panels beneath these cartoons. The relationship between these particular stages of illness development and the seven barriers to advancement is illustrated by the green panels (Figure 9).
The systematic study draws attention to the current shortcomings in the most advanced AI methods for brain tumor diagnosis. This literature review has focused on the challenges associated with the identification and assessment of brain tumors. The difficulties in detecting and diagnosing brain cancers early on are further compounded by the brain's intricate structure as an organ and their crucial role. Personalized treatment plans, the application of explainable AI in clinical decision making, and the integration of numerous data modalities are just a few of the topics that require further research and development. In general, solving these issues will speed up the creation of fully effective AI instruments for the clinical management of brain tumor patients.
None.
There is no conflicts of interest.

©2023 Ahmed, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.