MOJ
eISSN: 2379-6383


Research Article Special Issue Health Disorders
1Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Iran
2Infertility and Reproductive Health Research Center (IRHRC) and Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Iran
Correspondence: Farkhondeh Pouresmaeili, Infertility and Reproductive Health Research Center (IRHRC) and Department of Medical Genetics, Faculty of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Received: March 24, 2017 | Published: November 30, 2018
Citation: Aliniya N, Pouresmaeili F, Alan MS, et al. Maintaining the stability of placental rna in normal conditions. MOJ Public Health. 2018;7(6):304-305 DOI: 10.15406/mojph.2018.07.00255
For research on gene expression and its changes, maintenance of solid tissues like placenta is very important. Single–stranded RNA fragility under different environmental effects has prompted researchers to develop new RNA storage protocols to reduce entered damage to the tissue until extraction time. The applications of liquid nitrogen and RNA latter have been commonly used by investigators. We were looking for the simplest way to keep placental samples in appropriate condition with great RNA stability and function. Our experiments showed that storage of placental specimens in conventional freezing condition at –15 to –20 for short time and transfer at – 80 for longer time is as respondent as when applying liquid nitrogen or RNA later reagent.
Keywords: placenta, RNA later, RNA extraction
The handling, storage and work out with the excised tissue until extraction have been the investigators’ concern for years. The most essential requirement for gene expression study is the extraction of RNA in particular mRNA which is protein coding.1The RNA in total need to be protein free, intact, free of inhibitor enzymes which could interfere with the downstream segments of transcripts.2,3 Hence, the quality, purity and integrity of extracted RNAs beside the quantity are the values which are under improvement of developing kits for this purpose. Application of RNA latter (Qia– gen, www.qiagen.com)for tissue immersion and consequence storage at 4°C,–20°C and –80°C while in 1 mL of TRIzol Reagent (Invitrogen, www.invitrogen. com); also, flash–frozen in liquid nitrogen and storage at –80°C until tissue processing have been numerous methods to achieve an optimum way for preservation to keep the entire tissue nature and entity with no degradation in RNA property.4 We investigated the importance of storage procedure for solid tissue immediately after excision from the body.
Sample
Placental tissue samples were collected under sterile condition immediately after birth and transferred on ice and then to –20°C which latter on stored at –80°C. A breast tissue sample which was frozen in liquid nitrogen immediately after excision from the body was analyzed alongside with the placenta samples for B2M gene product analysis.
RNA extraction
We used 50mg of frozen tissue for total RNA isolation. Total RNA was extracted from placental tissues with TRizol reagent (Fisher Scientific Laboratories, USA) according to the manufacturer's protocol. In brief, the aqueous phase was resolved by addition of chloroform, and RNA precipitated from the aqueous phase by addition of isopropyl alcohol. Pelleted RNA was washed with 70% ethanol, dried, and re suspended in water.
cDNA synthesis
Reverse transcription was performed on one microgram of the recovered total RNA using Thermo Scientific Revert Aid First Strand cDNA Synthesis Kit (Fisher Scientific Laboratories, USA) according to the manufacturer's protocol. For this purpose, total RNA in 2µl was primed with 1µl of Random Hexamer primer and incubated for 5 min at 25°C followed by 60 min at 42°C. To investigate RNA quality, the expression of β2–microglobulin was assessed by RT–qPCR reaction.
Polymerase chain reaction
The PCR reaction for B2M gene amplification was performed in 20 μl volume containing 2µl of the synthesized cDNA, 10µl master mix, 0.5µl of each forward 5’–AGATGAGTATGCCTGCCGTG–3’ and reverse 5’–CGGCATCTTCAAACCTCCA–3’ primers and double distilled water was added up to 20µl (Ampliqon III, Denmark). PCR product was loaded on 2% agarose gel.
The produced cDNA was qualified on 2% agarose gel as is shown in the Figure 1. Two bands of 50 bp and 106 bp were observed where the frontier belonged to the primer dimers and the last one was related to the achieved PCR product of the target sample.
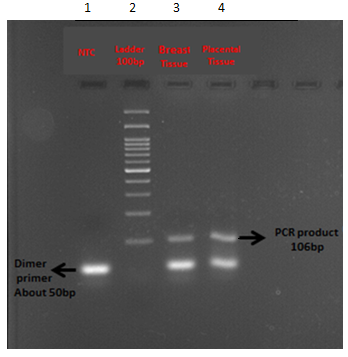
Figure 1Analysis of β2-microglobulin gene product on 2% agarose gel electrophoresis in two differently stored solid samples before RNA extraction.
Some investigations for functional genomics studies suggest freezing solid tissue samples in liquid nitrogen to stop all processes at a certain time and use for transcript mics.5,6 However, when the samples have to be shipped probably face to difficulties to be kept in frozen condition and the investigator does not know how long they will sit at the border where they might perish. RNA Later is probably a good choice for solid samples thicker than 5 mm.7–9 Otherwise we have to cut them. The storage temperature is very important. We can choose –80°C (or dry ice) removing the RNA Later after the first incubation or store at –20°C in the RNA later. The second method is the best to wait at the border, even at room temperature. However, this method prohibits temperature lower than –20°C otherwise you lose the RNA during defrost of the sample. Some researchers believe that for cell samples, RNA later leads to an important loss of RNA during centrifugation due to the viscosity of RNA Later even if the samples are treated by guanidine thiocianate and are transported on dry ice, Others grind the placenta tissue biopsies for RNA preservation and keep it in Trizol or similar reagent at –80ºC. As they suggest, RNA later is not a very good reagent when working with fungal material. Fungal cells are very tough and one needs mechanical force to break them. Somehow, RNA Later interferes with this decreasing the yield. My samples are extremely small so I need my RNA extraction to be as efficient as it can be. Here we showed no difference in the quantity and quality of the separated RNA from solid tissues either highly frozen or kept in an ordinary situation like 4°C and consequent –20°C. Moreover, our simple storage way is much cheaper than the other examined methods like usage of RNA latter or liquid nitrogen.
None.
The author declares no conflict of interest.

©2018 Aliniya, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.