MOJ
eISSN: 2379-6383


Review Article Volume 7 Issue 2
Pio XII College, Brazil
Correspondence: Rodrigo Pratte-Santos, Pio XII College, Bolívar de Abreu, 48 - Campo Grande, Cariacica - ES, Brazil, Tel +552733357365
Received: March 02, 2018 | Published: March 27, 2018
Citation: Mello MM, Vieira J, Simões FV, et al. Microbial resistance to colistin in consequence of mutations in themcr-1 gene of escherichia coli. MOJ Public Health. 2018;7(2):59 – 63. DOI: 10.15406/mojph.2018.07.00206
Objective: To evaluate the current situation of microbial resistance to colistin caused by mutations in the MCR-1 gene in Escherichia coli.
Materials and methods: A systematic review of the literature was carried out using scientific papers published in Scielo and Pubmed database since 2000.
Results: The plasmid-mediated colistin resistance MCR-1 gene is a transmissible gene that causes lipopolysaccharide modifications by regulation systems, presented on chronological scale to the inactivation of drugs that interact with the phospholipids and disorganize the structure of the cell membranes due to the indiscriminate use of colistin. Microbial resistance is mostly related to chromosomal mutations, however, when it refers to colistin it often occurs through horizontal gene transfer.
Conclusion: Whereas the mechanisms of microbial resistance to colistin it was possible to conclude that microbial resistance is mostly related to chromosomal mutations, however, when it refers to colistin it often occurs through horizontal gene transfer by plasmid.
Keywords: Escherichia coli, Colistin Resistance; MCR-1 gene
Have been shown around the world that Gram-negative bacteria are becoming resistant to many classes of antibiotics, representing a fearful situation for the onset of a future medical disaster. There are two terms used to describe this situation being experienced, which are Multi-Drug Resistant (MDR) and Pan-Drug Resistant (PDR). An isolate is considered MDR which exhibited resistance against 5 of the 7 anti-pseudomonal classes of Antimicrobial Agents, I.E. Anti-Pseudomonal Penicillins, Cephalosorins, Carbapenems, Monobactams, Quinolones, Aminoglycosides and colistin, and to be classified as a PDR be checked whether the isolates showed resistance to all anti-pseudomonal antimicrobial agents including colistin.1
The colistin (polymyxin E) has received great importance for treatments of last choice in infections caused by multiresistant Gram-negative bacteria.2 According to It was inserted into clinical practice in the mid 50's, but over the years its use was interrupted systematically with the growth of other active agents and Gram-negative bacteria, which were usually associated with manifestations of lower toxicity.3 The colistin is a polypeptide antibiotic and has been known for decades for combating Gram-negative bacteria, but in its composition there are substances that make it an antibiotic with nephrotoxic effects. Despite these effects, their use was reevaluated again because of its efficacy against multi-drug resistant Gram-negative bacteria. Colistin acts by inactivating the lipid A portion of the endotoxin present on the outer membrane of the bacterium, causing bacterial death.4
The resistance to this antibiotic in Gram-negative bacteria was associated with changes in the chromosome, specifically in pmrAB, phoPQ and mgrB genes, which affect lipid A of bacterial lipopolysaccharide.2 Have been found in isolates of Escherichia coli and Klebsiella pneumoniae of animal and human origin, a plasmidial transferable colistin resistance gene called MCR-1. The MCR-1 resistance gene was described in E. coli, K. pneumoniae, Enterobacter cloacae, among others. MCR-1 carriers were recovered in food of animal origin (swine, chickens, turkeys, cattle), plant, river water and in human patients. Furthermore, MCR-1 was found in strains isolated from these bacteria, but when it came into contact with extended-spectrum associated drugs such as beta-lactamases or KPC and carbapenemases, there was no resistance.5 Although the MCR-1 gene has recently been described as a resistance gene, it is widespread in Gram-negative bacteria in different countries of South America. In January 2016, the World Health Organization (WHO) shared with the Ministry of Health and Social Protection of Colombia a publication in which they report the detection of strains of Escherichia coli that carry the MCR-1 gene, which was found among travelers from Bolivia, Peru and Colombia.2 The finding of MCR-1-producing strains should be considered of high medical-epidemiological risk and requires the maximum effort of all members of health teams, especially the Infection Control Commission to avoid the spread of this type of resistance mechanism.5 The objective of this study was to evaluate the current situation of microbial resistance to colistin caused by mutations of the MCR-1 gene in Escherichia coli in high impact scientific literature.
A systematic literature review was performed on the Pubmed, Web of Science and Scielo database and on the keywords "Escherichia coli", "resistance", "MCR-1 gene" and "antibiotic". Performance of the MCR-1 resistance gene in Escherichia coli and on the antibiotic colistin. In this way, we were able to evaluate if there were changes in the resistance of this antibiotic and what the consequences on the inactivation of a antibiotic of last resort in public health. We approached the problem in a qualitative way, since it was necessary to analyze the action of the MCR-1 gene against the antibiotic colistin in each of the scenarios in a longitudinal study, in order to understand its effect on Escherichia coli. The data collection was done as illustrated in the following flowchart (Figure 1).
We selected articles published between 2000 and 2017, using Escherichia coli strains containing the MCR-1 gene, checking their mechanisms of action to make the bacteria resistant. We also used articles that report the action of the microorganism in humans, which contains the resistance gene that was submitted to the use of the antibiotic colistin to evaluate the alterations found. In order to compile our results, we have used studies published in scientific journals, that discuss the perspectives of antimicrobial pharmacology for the fight against microorganisms containing the MCR-1 resistance gene.
A total of 165 articles were identified in the period from 2000 to 2017. For the study, 78 articles were considered considering the criteria established in the methodology. We found 61 articles in Pubmed and 17 in Scielo. The articles selected come from countries where the situation of a post-antibiotic era approached more quickly, such as China and India, however, in this work articles were also used in which other countries such as the United States, Colombia, Brazil and Argentina reported incidences of the MCR-1 gene in Escherichia coli. And on the performance of the resistance gene, several studies that elucidate this mechanism in animals and foods were also used.
Microbial resistance to antimicrobials
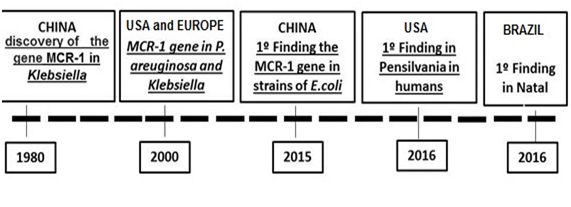
Resistance to antimicrobials is a genetic phenomenon, related to the existence of genes contained in the microorganism that encode different biochemical mechanisms that prevent the action of drugs. Resistance can originate from mutations that occur in the bacterium during its reproductive process and result from copy errors in the sequence of bases that form the chromosomal DNA, responsible for the genetic code. The other source of resistance is the import of the genes causing the phenomenon, consisting of transferable resistance. This resistance is done through the mechanisms of transduction, transformation and conjugation and often involves genes located in plasmids.6 In a retrospective study, researchers reported the identification of MCR-1 in three E. coli strains of chicken in China isolated in the 1980.7 As far as we know, this is the earliest identification of the MCR-1 gene reported in the scientific literature, however, there are reports of the discovery of the MCR-1 gene worldwide as illustrated in the following timeline (Figure 2). It has been observed that over time there is a great increase in the index of patients with E.coli carrying the MCR-1 gene and concomitant varied pathologies resulting from it. The problem that has been growing sharply and generating public health chaos is the indiscriminate use of antibiotics and chemotherapeutics, which has resulted in the development of bacterial species resistant to these drugs. Antibiotics, especially those composed of polypeptides, induce changes in permeability in the plasma membrane, changes that result in the loss of important metabolites by the microbial cell.7
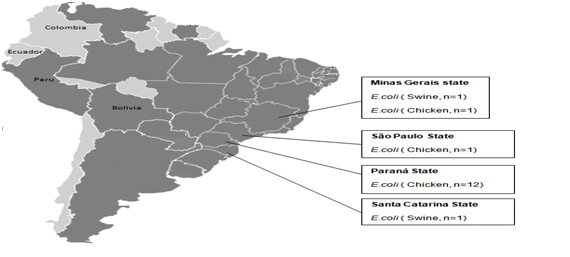
The recent discovery of the plasmid-mediated MCR-1 gene encoding colistin resistance in Escherichia coli and Klebsiella pneumoniae from animals, food and humans in China has initiated a search for this plasmid in different hosts and Gram-negative bacteria.8 The MCR-1 gene was identified on five continents in isolated bacteria of various origins including animals, foods, environments and humans.9,10 Yi-Liu and collaborators5 reported the first description of Plasmid-Mediated Colistin Resistance (MCR-1 gene) in animals, food, and humans in China. In Brazil, in a retrospective study of microbial resistance surveillance published in 2016,2 aimed to investigate between 2000 and 2016 resistance to colistin in 4,620 isolates of enterobacteria from animal, human, food and environmental samples. The MCR-1 gene was identified in 16 isolates from animal samples from the Southeast (Sao Paulo and Minas Gerais) and South (Parana and Santa Catarina) regions (Figure 3). Until 2016, this gene has been detected in three clinical specimens of E. coli, two of which come from patients attending hospitals in the city of São Paulo, Southeast Brazil, and one from a patient admitted to a hospital in the state of Rio Grande do Norte, Northern Brazil. There are also three samples of patients under analysis at the Adolfo Lutz Institute in São Paulo. It is important to note that all of these samples were sensitive to carbapenems and had low levels of resistance to polymyxins.2
The presence of E. coli with plasmid-mediated MCR-1 in Brazil is also relevant for the country's medical centers, where the emergence and spread of multiresistant pathogens, associated with high rates of treatment failure and use of colistin, intensive care units.11 The fact that the MCR-1 gene occurs in brazilian cattle is a matter of concern in terms of Brazil's overall contribution to the national and international movement of people and products, as this could contribute to accelerating the worldwide spread of the MCR-1. In fact, with a population of 205 million inhabitants, Brazil has continental proportions and is the largest country in Latin America. In addition, in agribusiness, it is the third producer of chicken meat (only after the United States and China) and the largest exporter of this product.2 An aggravating factor is that colistin sulfate is widely used in animal feed as a growth promoter in brazilian cattle, especially in swine and birds, supporting a link between the agricultural use of colistin and its resistance.
Current situation of microbial resistance to colistin
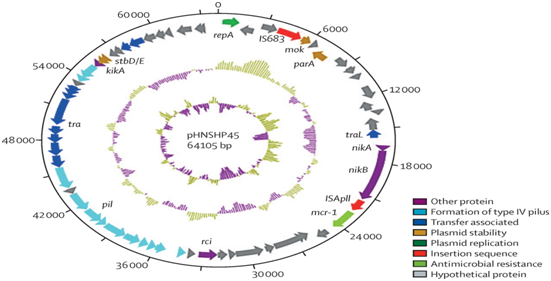
The resistance to colistin was related to chromosomal mutations and had never been reported to occur through horizontal gene transfer. During a routine surveillance project on antimicrobial resistance in commensal Escherichia coli from animals intended for consumption in China, a significant increase in resistance to colistin was observed. When a strain of E.coli, SHP45, resistant to colistin, which could have been transferred to another strain, was isolated from a pig, a more detailed analysis of the possible resistance to colistin mediated by plasmids was performed.5 Colistin resistance was found to be due only to the plasmid-mediated MCR-1 gene. The plasmid bearing MCR-1 was mobilized to an E. coli receptor at a frequency of 10-1 to 10-3 cells per receptor cell by conjugation and maintained in K. pneumoniae and Pseudomonas aeruginosa. In an in vivo model, the production of MCR-1 invalidated the efficacy of colistin. The MCR-1 gene is a member of the phosphoethanolaminatransferase enzyme family, being expressed in E. coli, resulting in the addition of phosphoethanolamine to lipid A, causing changes in the SHP45 plasmid (Figure 4).12
Temporal course of resistance and global reports
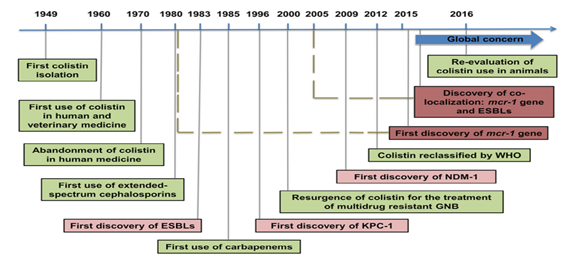
The emergence of MCR-1 heralds the violation of the last group of antibiotics, the polymyxins, by plasmid resistance. Currently, although more prevalent in China, MCR-1 is likely to match other global resistance. Mechanisms such as Metallo-Beta-Lactamase-1 (NDM-1), the findings of this study emphasize the urgent need for coordinated global action in the fight against pan-resistant Gram-negative bacteria. A retrospective study by Mohamed Rhouma et al.13 reported the identification of MCR-1 in strains of E. coli isolated in the 1980s, which seems to correspond to the first identification of ESBL enzymes. The first New Delhi Discovery (NDM-1) in 2009 was associated with a significant increase in the prevalence of the MCR-1 gene in strains of E. coli obtained from food and animal production. It has been observed that there is a historical link between MCR-1 and carbapenemase since 1980, it is believed that the re-evaluation of the use of colistin in animals needs an approach that includes not only the reduction of the use of colistin, but also the reduction of all use of antibiotics, to elucidate better, this fact is found in the following timeline (Figure 5).13
The MCR-1 gene is a polymyxin-resistant gene, and mainly polymyxin E, a drug considered the last resort for multiresistant bacteria treatments. The first gene report was in 2015 in food, animals and isolates from patients in China. The subject has been growing worldwide because it is the first mechanism of resistance to plasmid-mediated colistin. The major problem to be emphasized is the fact that the plasmids can be transferred between bacteria, allowing rapid dissemination. The MCR-1 gene was identified throughout the genome sequence of E. coli isolates in patients in the city of Connecticut. E. coli isolates were analyzed from feces collected from a patient with diarrhea. The patient had traveled to the Caribbean and spent approximately two weeks traveling. People who maintained contact with the patient were considered to be at risk for MCR-1 acquisition. In the investigation no transmission beyond the patient or persistent environmental contamination was identified, and the patient was transiently colonized.14
In other research, 980 strains of avian pathogenic Escherichia coli isolated from birds in the USA were examined. The MCR-1 gene was detected in 12 isolates recovered from diseased poultry from China and Egypt. MCR-1 positive isolates were also evaluated for resistance to phenotypic colistin and extended spectrum beta-lactam phenotypes and genotypes.15 In Thailand, one reports highlighted the appearance of E.coli carrying the drug-resistant MCR-1 gene. This study showed that the MCR-1 gene can be found among symptomatic and asymptomatic people, thus highlighting the need for a better understanding of the prevalence and epidemiology of this gene.16
Resistance mechanism of Escherichia coli to colistin
The mechanism of natural resistance of Gram-negative bacteria to polymyxins involves modifications of the outer lipopolysaccharide membrane through changes in lipid A. This results in the change in the negative charge of the bacterial cell surface and in the reduction of affinity with the positive charge of the antibiotic, as well making the interaction between the drug and the outer membrane of the cell difficult.17 These modifications are mediated by gene regulation systems, which are activated by environmental factors such as high concentrations of calcium and low concentrations of magnesium and also by changes in pH and the presence of iron.18
Gene regulation encompasses a two components system, which is used by several bacterial species to regulate the expression of different resistance factors as well as virulence. These components are constituted by a histidine kinase sensor protein that will undergo autophosphorylation when perceiving environmental stimuli, causing the activation of a second cytoplasmic protein, by a reaction of transphosphorylation. This second protein will then promote the activation or repression of target genes, thus triggering resistance to polymyxins.19 Colistin interacts electrostatically with the outer membrane of Gram-negative bacteria and migrates competitively with divalent cations that would stabilize the lipopolysaccharide layer, disrupting membrane integrity. Soon after, it is taken through the self-promoted uptake pathway. It is believed that colistin forms fissures in the outer membrane that promotes its uptake inside the cell allowing the passage of different molecules.20 Colistin is an amphipathic antibiotic, ie it has a hydrophilic region which is soluble in aqueous medium and also a hydrophobic region, ie insoluble in water, but soluble in lipids and organic solvent. Colistin acts primarily on the outer and cytoplasmic membranes of the bacterium, having a similar action as that of the cationic detergent. The colistin binds to cellular envelope components, such as phospholipids and lipopolysaccharides, causing the displacement of Ca and Mg ions that act as membrane stabilizers, causing the rupture thereof, leading to loss of the cellular content and causing the death of the bacterium.21
In general, in colistin, two phenotypes that create resistance are observed. The first is the natural resistance, possibly resulting from mutations in the bacterial genome, promoting resistance with MICs close to the cut-off points of the resistance category. The second phenotype is called the adaptive mechanism, when the initially sensitive bacterium becomes resistant after prolonged use of the drug in sub-optimal dosages. The adaptive mechanism may lead to much higher MICs above 128 ug/mL, and this phenotype is determined reversible in the absence of drug-selective pressure.19 In Brazil, was used colistin to treat infection in 109 patients. The high mortality rate after the start of use (71.6% of the patients) is highlighted. Twenty-nine patients (26.6%) developed renal lesions prior to treatment with colistin. Only six of them recovered renal function during treatment with colistin. Twenty-one patients developed acute renal injury, 11 of them requiring dialysis treatment. The use of vasopressors were the factors that led the patients to death, according to the logistic regression model.22 In Thailand, was studied seventy-eight patients with ages of approximately 63.5 years. Nephrotoxicity was observed in at least twenty-four patients (30.8%), and seventeen showed other predisposing factors for renal damage (other nephrotoxic drugs, chronic renal disease and hypovolemia). Renal impairment was considered mild and reversible, with no need for dialysis therapy. Mortality at 30 days was 46%.23
In USAwas evaluated 4,438 E. coli isolates of animal origin. The susceptibility to colistin was tested by the agar dilution method. The mutations in pmrA, pmrB and mgrB and the presence of the MCR-1 gene were determined by PCR and DNA sequencing. Complementation experiments were performed to evaluate the contribution of mutations to resistance to colistin. There was a high prevalence of resistance to colistin in farm E. coli (24.1%) and slaughter (24.3%) in 2013-2014, followed by chickens on the farm (14.0%) and slaughter (9.5%), however, 91.0% of isolates resistant to colistin were positive for MCR-1. The high frequency of colistin resistance and the MCR-1 gene among E. coli isolates from food animals in China called for the need to minimize the potential risks of developing colistin resistance and the spread of the MCR-1 gene.24 The extensive use of colistin in veterinary medicine to treat gastrointestinal infections caused by Enterobacteriaceae has contributed to MCR-1 chromosomal mutations mediated by plasmid C to encode resistance to colistin in Gram-negative.25
Microbial resistance is mostly related to chromosomal mutations, however, when it refers to colistin it often occurs through horizontal gene transfer. Regarding the mechanisms of microbial resistance to colistin, it was possible to conclude that in Escherichia coli and Klebsiella pneumoniae bacteria there is a prevalence of the colistin resistant SHP45 plasmid, which carries the MCR-1 gene.With respect to the resistance of Escherichia coli to colistin, it has been possible to analyze that colistin is a potent antibiotic that acts on the outer membrane of Gram-negative bacteria; however, in mutations in the pmrA gene, pmrB and mgrB were the presence of the MCR-1 gene, which inactivates the efficacy of this antibiotic. Finally, global reports of microbial resistance to colistin showed that the first findings on MCR-1 gene occurred in chickens in the year 1980 in China and over the years the presence of this resistance gene in Gram-negative bacteria has become more frequent, being spread on five continents in isolates of food, animals and even in humans.
To the Scientific Initiation Program of the School of Biomedical Sciences of Espirito Santo (PIO XII College).
The authors declare that they have no conflict of interests.

©2018 Mello, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.